Nitrogen fixation and nitrogenase (nifH) expression in tropical waters of the eastern North Atlantic
- PMID: 21228888
- PMCID: PMC3146282
- DOI: 10.1038/ismej.2010.205
Nitrogen fixation and nitrogenase (nifH) expression in tropical waters of the eastern North Atlantic
Abstract
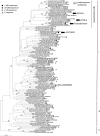
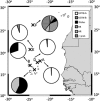
Expression of nifH in 28 surface water samples collected during fall 2007 from six stations in the vicinity of the Cape Verde Islands (north-east Atlantic) was examined using reverse transcription-polymerase chain reaction (RT-PCR)-based clone libraries and quantitative RT-PCR (RT-qPCR) analysis of seven diazotrophic phylotypes. Biological nitrogen fixation (BNF) rates and nutrient concentrations were determined for these stations, which were selected based on a range in surface chlorophyll concentrations to target a gradient of primary productivity. BNF rates greater than 6 nmolN l(-1) h(-1) were measured at two of the near-shore stations where high concentrations of Fe and PO(4)(3-) were also measured. Six hundred and five nifH transcripts were amplified by RT-PCR, of which 76% are described by six operational taxonomic units, including Trichodesmium and the uncultivated UCYN-A, and four non-cyanobacterial diazotrophs that clustered with uncultivated Proteobacteria. Although all five cyanobacterial phylotypes quantified in RT-qPCR assays were detected at different stations in this study, UCYN-A contributed most significantly to the pool of nifH transcripts in both coastal and oligotrophic waters. A comparison of results from RT-PCR clone libraries and RT-qPCR indicated that a γ-proteobacterial phylotype was preferentially amplified in clone libraries, which underscores the need to use caution interpreting clone-library-based nifH studies, especially when considering the importance of uncultivated proteobacterial diazotrophs.
Figures

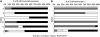
References
-
- Bergman B, Gallon JR, Rai AN, Stal LJ. N2 fixation by non-heterocystous cyanobacteria. FEMS Microbiol Rev. 1997;19:139–185.
-
- Berman-Frank I, Quigg A, Finkel ZV, Irwin AJ, Haramaty L. Nitrogen-fixation strategies and Fe requirements in cyanobacteria. Limnol Oceanogr. 2007;52:2260–2269.
-
- Brewer PG, Riley JP. The automatic determination of nitrate in sea water. Deep-Sea Res. 1965;12:765–772.

