Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis
- PMID: 21232103
- PMCID: PMC3091300
- DOI: 10.1186/gb-2011-12-1-r2
Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis
Abstract
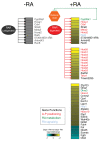
Background: Among its many roles in development, retinoic acid determines the anterior-posterior identity of differentiating motor neurons by activating retinoic acid receptor (RAR)-mediated transcription. RAR is thought to bind the genome constitutively, and only induce transcription in the presence of the retinoid ligand. However, little is known about where RAR binds to the genome or how it selects target sites.
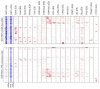
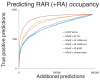
Results: We tested the constitutive RAR binding model using the retinoic acid-driven differentiation of mouse embryonic stem cells into differentiated motor neurons. We find that retinoic acid treatment results in widespread changes in RAR genomic binding, including novel binding to genes directly responsible for anterior-posterior specification, as well as the subsequent recruitment of the basal polymerase machinery. Finally, we discovered that the binding of transcription factors at the embryonic stem cell stage can accurately predict where in the genome RAR binds after initial differentiation.
Conclusions: We have characterized a ligand-dependent shift in RAR genomic occupancy at the initiation of neurogenesis. Our data also suggest that enhancers active in pluripotent embryonic stem cells may be preselecting regions that will be activated by RAR during neuronal differentiation.
Figures



Similar articles
-
RAR/RXR binding dynamics distinguish pluripotency from differentiation associated cis-regulatory elements.Nucleic Acids Res. 2015 May 26;43(10):4833-54. doi: 10.1093/nar/gkv370. Epub 2015 Apr 20. Nucleic Acids Res. 2015. PMID: 25897113 Free PMC article.
-
Retinoid X receptor (RXR) within the RXR-retinoic acid receptor heterodimer binds its ligand and enhances retinoid-dependent gene expression.Mol Cell Biol. 1997 Feb;17(2):644-55. doi: 10.1128/MCB.17.2.644. Mol Cell Biol. 1997. PMID: 9001218 Free PMC article.
-
Chromosomal integration of retinoic acid response elements prevents cooperative transcriptional activation by retinoic acid receptor and retinoid X receptor.Mol Cell Biol. 2002 Mar;22(5):1446-59. doi: 10.1128/MCB.22.5.1446-1459.2002. Mol Cell Biol. 2002. PMID: 11839811 Free PMC article.
-
Retinoic acid signaling and mouse embryonic stem cell differentiation: Cross talk between genomic and non-genomic effects of RA.Biochim Biophys Acta. 2015 Jan;1851(1):66-75. doi: 10.1016/j.bbalip.2014.04.003. Epub 2014 Apr 24. Biochim Biophys Acta. 2015. PMID: 24768681 Review.
-
The retinoid receptors.Leukemia. 1994 Nov;8(11):1797-806. Leukemia. 1994. PMID: 7967725 Review.
Cited by
-
The elongation complex components BRD4 and MLLT3/AF9 are transcriptional coactivators of nuclear retinoid receptors.PLoS One. 2013 Jun 10;8(6):e64880. doi: 10.1371/journal.pone.0064880. Print 2013. PLoS One. 2013. PMID: 23762261 Free PMC article.
-
RAR/RXR binding dynamics distinguish pluripotency from differentiation associated cis-regulatory elements.Nucleic Acids Res. 2015 May 26;43(10):4833-54. doi: 10.1093/nar/gkv370. Epub 2015 Apr 20. Nucleic Acids Res. 2015. PMID: 25897113 Free PMC article.
-
HDAC1-mediated repression of the retinoic acid-responsive gene ripply3 promotes second heart field development.PLoS Genet. 2019 May 15;15(5):e1008165. doi: 10.1371/journal.pgen.1008165. eCollection 2019 May. PLoS Genet. 2019. PMID: 31091225 Free PMC article.
-
Global Transcriptional Analyses of the Wnt-Induced Development of Neural Stem Cells from Human Pluripotent Stem Cells.Int J Mol Sci. 2021 Jul 12;22(14):7473. doi: 10.3390/ijms22147473. Int J Mol Sci. 2021. PMID: 34299091 Free PMC article.
-
Induction of specific neuron types by overexpression of single transcription factors.In Vitro Cell Dev Biol Anim. 2016 Oct;52(9):961-973. doi: 10.1007/s11626-016-0056-7. Epub 2016 Jun 1. In Vitro Cell Dev Biol Anim. 2016. PMID: 27251161 Free PMC article.
References
-
- Corral RDD, Storey KG. Opposing FGF and retinoid pathways: a signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. BioEssays. 2004;26:857–869. - PubMed
-
- Niederreither K, Vermot J, Schuhbaur B, Chambon P, Dollé P. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development. 2000;127:75–85. - PubMed
-
- Liu JP, Laufer E, Jessell TM. Assigning the positional identity of spinal motor neurons: rostrocaudal patterning of Hox-c expression by FGFs, Gdf11, and retinoids. Neuron. 2001;32:997–1012. - PubMed
-
- Sockanathan S, Jessell TM. Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell. 1998;94:503–514. - PubMed
-
- Novitch B, Wichterle H, Jessell T, Sockanathan S. A requirement for retinoic acid-mediated transcriptional activation in ventral neural patterning and motor neuron specification. Neuron. 2003;40:81–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

