Genomic fluidity: an integrative view of gene diversity within microbial populations
- PMID: 21232151
- PMCID: PMC3030549
- DOI: 10.1186/1471-2164-12-32
Genomic fluidity: an integrative view of gene diversity within microbial populations
Abstract
Background: The dual concepts of pan and core genomes have been widely adopted as means to assess the distribution of gene families within microbial species and genera. The core genome is the set of genes shared by a group of organisms; the pan genome is the set of all genes seen in any of these organisms. A variety of methods have provided drastically different estimates of the sizes of pan and core genomes from sequenced representatives of the same groups of bacteria.
Results: We use a combination of mathematical, statistical and computational methods to show that current predictions of pan and core genome sizes may have no correspondence to true values. Pan and core genome size estimates are problematic because they depend on the estimation of the occurrence of rare genes and genomes, respectively, which are difficult to estimate precisely because they are rare. Instead, we introduce and evaluate a robust metric - genomic fluidity - to categorize the gene-level similarity among groups of sequenced isolates. Genomic fluidity is a measure of the dissimilarity of genomes evaluated at the gene level.
Conclusions: The genomic fluidity of a population can be estimated accurately given a small number of sequenced genomes. Further, the genomic fluidity of groups of organisms can be compared robustly despite variation in algorithms used to identify genes and their homologs. As such, we recommend that genomic fluidity be used in place of pan and core genome size estimates when assessing gene diversity within genomes of a species or a group of closely related organisms.
Figures

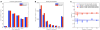

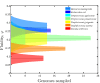
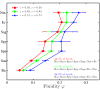

References
-
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24(3):133–141. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

