Exploration of the cytochrome c oxidase pathway puzzle and examination of the origin of elusive mutational effects
- PMID: 21232525
- PMCID: PMC3055932
- DOI: 10.1016/j.bbabio.2011.01.004
Exploration of the cytochrome c oxidase pathway puzzle and examination of the origin of elusive mutational effects
Abstract
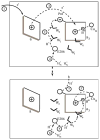
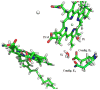
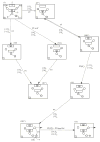
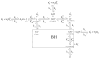
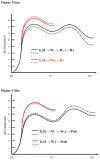
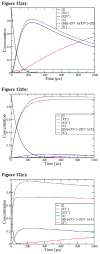
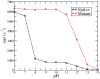
Gaining detailed understanding of the energetics of the proton-pumping process in cytochrome c oxidase (CcO) is a problem of great current interest. Despite promising mechanistic proposals, so far, a physically consistent model that would reproduce all the relevant barriers needed to create a working pump has not been presented. In addition, there are major problems in elucidating the origin of key mutational effects and in understanding the nature of the apparent pK(a) values associated with the pH dependencies of specific proton transfer (PT) reactions in CcO. This work takes a key step in resolving the above problems, by considering mutations, such as the Asn139Asp replacement, that blocks proton pumping without affecting PT to the catalytic site. We first introduce a formulation that makes it possible to relate the apparent pK(a) of Glu286 to different conformational states of this residue. We then use the new formulation along with the calculated pK(a) values of Glu286 at these different conformations to reproduce the experimentally observed apparent pK(a) of the residue. Next, we take the X-ray structures of the native and Asn139Asp mutant of the Paracoccus denitrificans CcO (N131D in this system) and reproduce for the first time the change in the primary PT pathways (and other key features) based on simulations that start with the observed structural changes. We also consider the competition between proton transport to the catalytic site and the pump site, as a function of the bulk pH, as well as the H/D isotope effect, and use this information to explore the relative height of the two barriers. The paper emphasizes the crucial role of energy-based considerations that include the PT process, and the delicate control of PT in CcO.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures

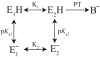




Similar articles
-
Exploring pathways and barriers for coupled ET/PT in cytochrome c oxidase: a general framework for examining energetics and mechanistic alternatives.Biochim Biophys Acta. 2007 Mar;1767(3):244-60. doi: 10.1016/j.bbabio.2007.01.015. Epub 2007 Jan 30. Biochim Biophys Acta. 2007. PMID: 17350588 Free PMC article.
-
A D-pathway mutation decouples the Paracoccus denitrificans cytochrome c oxidase by altering the side-chain orientation of a distant conserved glutamate.J Mol Biol. 2008 Dec 26;384(4):865-77. doi: 10.1016/j.jmb.2008.09.074. Epub 2008 Oct 9. J Mol Biol. 2008. PMID: 18930738
-
Characterizing the proton loading site in cytochrome c oxidase.Proc Natl Acad Sci U S A. 2014 Aug 26;111(34):12414-9. doi: 10.1073/pnas.1407187111. Epub 2014 Aug 11. Proc Natl Acad Sci U S A. 2014. PMID: 25114210 Free PMC article.
-
Respiratory conservation of energy with dioxygen: cytochrome C oxidase.Met Ions Life Sci. 2015;15:89-130. doi: 10.1007/978-3-319-12415-5_4. Met Ions Life Sci. 2015. PMID: 25707467 Review.
-
Cytochrome c oxidase (heme aa3) from Paracoccus denitrificans: analysis of mutations in putative proton channels of subunit I.J Bioenerg Biomembr. 1998 Feb;30(1):89-97. doi: 10.1023/a:1020515713103. J Bioenerg Biomembr. 1998. PMID: 9623810 Review.
Cited by
-
Mutation of a single residue in the ba3 oxidase specifically impairs protonation of the pump site.Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):3397-402. doi: 10.1073/pnas.1422434112. Epub 2015 Mar 2. Proc Natl Acad Sci U S A. 2015. PMID: 25733886 Free PMC article.
-
On the control of the proton current in the voltage-gated proton channel Hv1.Proc Natl Acad Sci U S A. 2018 Oct 9;115(41):10321-10326. doi: 10.1073/pnas.1809766115. Epub 2018 Sep 25. Proc Natl Acad Sci U S A. 2018. PMID: 30254162 Free PMC article.
-
Cavity hydration dynamics in cytochrome c oxidase and functional implications.Proc Natl Acad Sci U S A. 2017 Oct 17;114(42):E8830-E8836. doi: 10.1073/pnas.1707922114. Epub 2017 Oct 2. Proc Natl Acad Sci U S A. 2017. PMID: 28973914 Free PMC article.
-
Analyzing the electrogenicity of cytochrome c oxidase.Proc Natl Acad Sci U S A. 2016 Jul 12;113(28):7810-5. doi: 10.1073/pnas.1608118113. Epub 2016 Jun 28. Proc Natl Acad Sci U S A. 2016. PMID: 27357681 Free PMC article.
-
Proton-transport mechanisms in cytochrome c oxidase revealed by studies of kinetic isotope effects.Biochim Biophys Acta. 2011 Sep;1807(9):1083-94. doi: 10.1016/j.bbabio.2011.03.012. Epub 2011 Apr 2. Biochim Biophys Acta. 2011. PMID: 21463601 Free PMC article.
References
-
- Wikström MKF. Proton pump coupled to cytochrome-c oxidase in mitochondria. Nature. 1977;266(5599):271–273. - PubMed
-
- Michel H, et al. Cytochrome c oxidase: structure and spectroscopy. Annu Rev Biophys Biomol Struct. 1998;27:329–56. - PubMed
-
- Brzezinski P, Larsson G. Redox-driven proton pumping by heme-copper oxidases. Biochim Biophys Acta - Bioenergetics. 2003;1605(1–3):1–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

