High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy
- PMID: 21233418
- PMCID: PMC3033257
- DOI: 10.1073/pnas.1018157108
High-throughput ectopic expression screen for tamoxifen resistance identifies an atypical kinase that blocks autophagy
Abstract
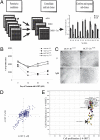
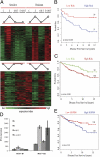
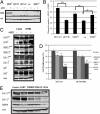
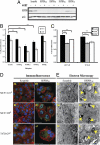
Resistance to tamoxifen in breast cancer patients is a serious therapeutic problem and major efforts are underway to understand underlying mechanisms. Resistance can be either intrinsic or acquired. We derived a series of subcloned MCF7 cell lines that were either highly sensitive or naturally resistant to tamoxifen and studied the factors that lead to drug resistance. Gene-expression studies revealed a signature of 67 genes that differentially respond to tamoxifen in sensitive vs. resistant subclones, which also predicts disease-free survival in tamoxifen-treated patients. High-throughput cell-based screens, in which >500 human kinases were independently ectopically expressed, identified 31 kinases that conferred drug resistance on sensitive cells. One of these, HSPB8, was also in the expression signature and, by itself, predicted poor clinical outcome in one cohort of patients. Further studies revealed that HSPB8 protected MCF7 cells from tamoxifen and blocked autophagy. Moreover, silencing HSBP8 induced autophagy and caused cell death. Tamoxifen itself induced autophagy in sensitive cells but not in resistant ones, and tamoxifen-resistant cells were sensitive to the induction of autophagy by other drugs. These results may point to an important role for autophagy in the sensitivity to tamoxifen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. - PubMed
-
- Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer. 2004;11:643–658. - PubMed
-
- Frasor J, et al. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 2006;66:7334–7340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

