Epigenetic regulation of vascular smooth muscle cell proliferation and neointima formation by histone deacetylase inhibition
- PMID: 21233448
- PMCID: PMC3074344
- DOI: 10.1161/ATVBAHA.110.221952
Epigenetic regulation of vascular smooth muscle cell proliferation and neointima formation by histone deacetylase inhibition
Abstract
Objective: Proliferation of smooth muscle cells (SMC) in response to vascular injury is central to neointimal vascular remodeling. There is accumulating evidence that histone acetylation constitutes a major epigenetic modification for the transcriptional control of proliferative gene expression; however, the physiological role of histone acetylation for proliferative vascular disease remains elusive.
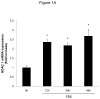
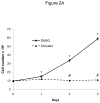
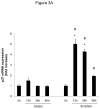
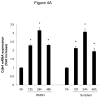
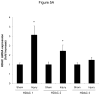
Methods and results: In the present study, we investigated the role of histone deacetylase (HDAC) inhibition in SMC proliferation and neointimal remodeling. We demonstrate that mitogens induce transcription of HDAC 1, 2, and 3 in SMC. Short interfering RNA-mediated knockdown of either HDAC 1, 2, or 3 and pharmacological inhibition of HDAC prevented mitogen-induced SMC proliferation. The mechanisms underlying this reduction of SMC proliferation by HDAC inhibition involve a growth arrest in the G(1) phase of the cell cycle that is due to an inhibition of retinoblastoma protein phosphorylation. HDAC inhibition resulted in a transcriptional and posttranscriptional regulation of the cyclin-dependent kinase inhibitors p21(Cip1) and p27(Kip). Furthermore, HDAC inhibition repressed mitogen-induced cyclin D1 mRNA expression and cyclin D1 promoter activity. As a result of this differential cell cycle-regulatory gene expression by HDAC inhibition, the retinoblastoma protein retains a transcriptional repression of its downstream target genes required for S phase entry. Finally, we provide evidence that these observations are applicable in vivo by demonstrating that HDAC inhibition decreased neointima formation and expression of cyclin D1 in a murine model of vascular injury.
Conclusions: These findings identify HDAC as a critical component of a transcriptional cascade regulating SMC proliferation and suggest that HDAC might play a pivotal role in the development of proliferative vascular diseases, including atherosclerosis and in-stent restenosis.
Conflict of interest statement
Disclosure - The authors have nothing to disclose that could be perceived as real or apparent conflict(s) of interest.
Figures


















Comment in
-
Drugs targeting epigenetic histone acetylation in vascular smooth muscle cells for restenosis and atherosclerosis.Arterioscler Thromb Vasc Biol. 2011 Apr;31(4):725-7. doi: 10.1161/ATVBAHA.111.222976. Arterioscler Thromb Vasc Biol. 2011. PMID: 21411753 Free PMC article. No abstract available.
References
-
- Dzau VJ, Braun-Dullaeus RC, Sedding DG. Vascular proliferation and atherosclerosis: New perspectives and therapeutic strategies. Nat Med. 2002;8:1249–1256. - PubMed
-
- Moses JW, Leon MB, Popma JJ, Fitzgerald PJ, Holmes DR, O’Shaughnessy C, Caputo RP, Kereiakes DJ, Williams DO, Teirstein PS, Jaeger JL, Kuntz RE the SIRIUS Investigators. Sirolimus-eluting stents versus standard stents in patients with stenosis in a native coronary artery. N Engl J Med. 2003;349:1315–1323. - PubMed
-
- Zhang QJ, Goddard M, Shanahan C, Shapiro L, Bennett M. Differential gene expression in vascular smooth muscle cells in primary atherosclerosis and in stent stenosis in humans. Arterioscler Thromb Vasc Biol. 2002;22:2030–2036. - PubMed
-
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

