The genome sequence of Methanohalophilus mahii SLP(T) reveals differences in the energy metabolism among members of the Methanosarcinaceae inhabiting freshwater and saline environments
- PMID: 21234345
- PMCID: PMC3017947
- DOI: 10.1155/2010/690737
The genome sequence of Methanohalophilus mahii SLP(T) reveals differences in the energy metabolism among members of the Methanosarcinaceae inhabiting freshwater and saline environments
Abstract
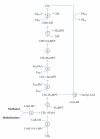
Methanohalophilus mahii is the type species of the genus Methanohalophilus, which currently comprises three distinct species with validly published names. Mhp. mahii represents moderately halophilic methanogenic archaea with a strictly methylotrophic metabolism. The type strain SLP(T) was isolated from hypersaline sediments collected from the southern arm of Great Salt Lake, Utah. Here we describe the features of this organism, together with the complete genome sequence and annotation. The 2,012,424 bp genome is a single replicon with 2032 protein-coding and 63 RNA genes and part of the Genomic Encyclopedia of Bacteria and Archaea project. A comparison of the reconstructed energy metabolism in the halophilic species Mhp. mahii with other representatives of the Methanosarcinaceae reveals some interesting differences to freshwater species.
Figures





References
-
- Paterek JR, Smith PH. Methanophilus mahii gen. nov., sp. nov., a methylotrophic halophilic methanogen. International Journal of Systematic Bacteriology. 1988;38(1):122–123.
-
- Chun J, Lee JH, Jung Y, et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. International Journal of Systematic and Evolutionary Microbiology. 2007;57(10):2259–2261. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions

