Quality control procedures for genome-wide association studies
- PMID: 21234875
- PMCID: PMC3066182
- DOI: 10.1002/0471142905.hg0119s68
Quality control procedures for genome-wide association studies
Abstract
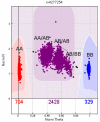
Genome-wide association studies (GWAS) are being conducted at an unprecedented rate in population-based cohorts and have increased our understanding of the pathophysiology of complex disease. Regardless of context, the practical utility of this information will ultimately depend upon the quality of the original data. Quality control (QC) procedures for GWAS are computationally intensive, operationally challenging, and constantly evolving. Here we enumerate some of the challenges in QC of GWAS data and describe the approaches that the electronic MEdical Records and Genomics (eMERGE) network is using for quality assurance in GWAS data, thereby minimizing potential bias and error in GWAS results. We discuss common issues associated with QC of GWAS data, including data file formats, software packages for data manipulation and analysis, sex chromosome anomalies, sample identity, sample relatedness, population substructure, batch effects, and marker quality. We propose best practices and discuss areas of ongoing and future research.
© 2011 by John Wiley & Sons, Inc.
Figures





References
-
- Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8:657–662. - PubMed
Publication types
MeSH terms
Grants and funding
- U01HG004438/HG/NHGRI NIH HHS/United States
- T32 GM080178/GM/NIGMS NIH HHS/United States
- U01HG004610/HG/NHGRI NIH HHS/United States
- U01 HG004603/HG/NHGRI NIH HHS/United States
- U01HG04603/HG/NHGRI NIH HHS/United States
- U01HG004609/HG/NHGRI NIH HHS/United States
- U01 HG004609/HG/NHGRI NIH HHS/United States
- U01 HG004599/HG/NHGRI NIH HHS/United States
- U01HG004608/HG/NHGRI NIH HHS/United States
- U01 HG004608/HG/NHGRI NIH HHS/United States
- U01 HG006375/HG/NHGRI NIH HHS/United States
- U01 HG004438/HG/NHGRI NIH HHS/United States
- U01HG04599/HG/NHGRI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 LM010040/LM/NLM NIH HHS/United States
- U01 HG004610/HG/NHGRI NIH HHS/United States
- R01LM010040/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

