Full mitochondrial genome sequences of two endemic Philippine hornbill species (Aves: Bucerotidae) provide evidence for pervasive mitochondrial DNA recombination
- PMID: 21235758
- PMCID: PMC3025957
- DOI: 10.1186/1471-2164-12-35
Full mitochondrial genome sequences of two endemic Philippine hornbill species (Aves: Bucerotidae) provide evidence for pervasive mitochondrial DNA recombination
Abstract
Background: Although nowaday it is broadly accepted that mitochondrial DNA (mtDNA) may undergo recombination, the frequency of such recombination remains controversial. Its estimation is not straightforward, as recombination under homoplasmy (i.e., among identical mt genomes) is likely to be overlooked. In species with tandem duplications of large mtDNA fragments the detection of recombination can be facilitated, as it can lead to gene conversion among duplicates. Although the mechanisms for concerted evolution in mtDNA are not fully understood yet, recombination rates have been estimated from "one per speciation event" down to 850 years or even "during every replication cycle".
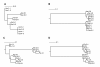
Results: Here we present the first complete mt genome of the avian family Bucerotidae, i.e., that of two Philippine hornbills, Aceros waldeni and Penelopides panini. The mt genomes are characterized by a tandemly duplicated region encompassing part of cytochrome b, 3 tRNAs, NADH6, and the control region. The duplicated fragments are identical to each other except for a short section in domain I and for the length of repeat motifs in domain III of the control region. Due to the heteroplasmy with regard to the number of these repeat motifs, there is some size variation in both genomes; with around 21,657 bp (A. waldeni) and 22,737 bp (P. panini), they significantly exceed the hitherto longest known avian mt genomes, that of the albatrosses. We discovered concerted evolution between the duplicated fragments within individuals. The existence of differences between individuals in coding genes as well as in the control region, which are maintained between duplicates, indicates that recombination apparently occurs frequently, i.e., in every generation.
Conclusions: The homogenised duplicates are interspersed by a short fragment which shows no sign of recombination. We hypothesize that this region corresponds to the so-called Replication Fork Barrier (RFB), which has been described from the chicken mitochondrial genome. As this RFB is supposed to halt replication, it offers a potential mechanistic explanation for frequent recombination in mitochondrial genomes.
Figures



References
-
- Eberhard JR, Wright TF, Bermingham E. Duplication and concerted evolution of the mitochondrial control region in the parrot genus Amazona. Mol Biol Evol. 2001;18:1330–1342. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

