Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism
- PMID: 21239497
- PMCID: PMC3048721
- DOI: 10.1074/jbc.M110.203380
Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism
Abstract
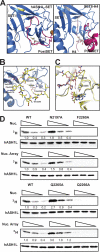
Absent, small, or homeotic disc1 (Ash1) is a trithorax group histone methyltransferase that is involved in gene activation. Although there are many known histone methyltransferases, their regulatory mechanisms are poorly understood. Here, we present the crystal structure of the human ASH1L catalytic domain, showing its substrate binding pocket blocked by a loop from the post-SET domain. In this configuration, the loop limits substrate access to the active site. Mutagenesis of the loop stimulates ASH1L histone methyltransferase activity, suggesting that ASH1L activity may be regulated through the loop from the post-SET domain. In addition, we show that human ASH1L specifically methylates histone H3 Lys-36. Our data implicate that there may be a regulatory mechanism of ASH1L histone methyltransferases.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

