Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+
- PMID: 21239683
- PMCID: PMC3033254
- DOI: 10.1073/pnas.1017659108
Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+
Abstract
Trinitrophenyl derivatives of adenine nucleotides are widely used for probing ATP-binding sites. Here we describe crystal structures of Ca(2+)-ATPase, a representative P-type ATPase, in the absence of Ca(2+) with bound ATP, trinitrophenyl-ATP, -ADP, and -AMP at better than 2.4-Å resolution, stabilized with thapsigargin, a potent inhibitor. These crystal structures show that the binding mode of the trinitrophenyl derivatives is distinctly different from the parent adenine nucleotides. The adenine binding pocket in the nucleotide binding domain of Ca(2+)-ATPase is now occupied by the trinitrophenyl group, and the side chains of two arginines sandwich the adenine ring, accounting for the much higher affinities of the trinitrophenyl derivatives. Trinitrophenyl nucleotides exhibit a pronounced fluorescence in the E2P ground state but not in the other E2 states. Crystal structures of the E2P and E2 ∼ P analogues of Ca(2+)-ATPase with bound trinitrophenyl-AMP show that different arrangements of the three cytoplasmic domains alter the orientation and water accessibility of the trinitrophenyl group, explaining the origin of "superfluorescence." Thus, the crystal structures demonstrate that ATP and its derivatives are highly adaptable to a wide range of site topologies stabilized by a variety of interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

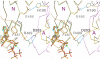

 , an E2 ∼ P transition state analogue, and (C)
, an E2 ∼ P transition state analogue, and (C)  , an E2P ground state analogue. The A, N, and P domains are colored pink, green, and yellow, respectively. Blue broken lines show hydrogen bonds. Small red spheres represent water molecules; small cyan disks, main chain amide. Carbon atoms in the TNP group are colored gray.
, an E2P ground state analogue. The A, N, and P domains are colored pink, green, and yellow, respectively. Blue broken lines show hydrogen bonds. Small red spheres represent water molecules; small cyan disks, main chain amide. Carbon atoms in the TNP group are colored gray.
 (B), and
(B), and  (C). Prepared with PyMol (40). A stereo version of this figure is presented as
(C). Prepared with PyMol (40). A stereo version of this figure is presented as References
-
- Hiratsuka T, Uchida K. Preparation and properties of 2′(or 3′)-O-(2,4,6-trinitrophenyl) adenosine 5′-triphosphate, an analog of adenosine triphosphate. Biochim Biophys Acta. 1973;320:635–647. - PubMed
-
- Watanabe T, Inesi G. The use of 2′,3′-O-(2,4,6-trinitrophenyl) adenosine 5′-triphosphate for studies of nucleotide interaction with sarcoplasmic reticulum vesicles. J Biol Chem. 1982;257:11510–11516. - PubMed
-
- Dupont Y, Chapron Y, Pougeois R. Titration of the nucleotide binding sites of sarcoplasmic reticulum Ca2+-ATPase with 2′,3′-O-(2,4,6-trinitrophenyl) adenosine 5′-triphosphate and 5′-diphosphate. Biochem Biophys Res Commun. 1982;106:1272–1279. - PubMed
-
- Moczydlowski EG, Fortes PA. Characterization of 2′,3′-O-(2,4,6-trinitrocyclohexadienylidine)adenosine 5′-triphosphate as a fluorescent probe of the ATP site of sodium and potassium transport adenosine triphosphatase. J Biol Chem. 1981;256:2346–2356. - PubMed
-
- Seebregts CJ, McIntosh DB. 2′,3′-O-(2,4,6-trinitrophenyl)-8-azido-adenosine mono-, di-, and triphosphates as photoaffinity probes of the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem. 1989;264:2043–2052. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

