Structure of UvrA nucleotide excision repair protein in complex with modified DNA
- PMID: 21240268
- PMCID: PMC3428727
- DOI: 10.1038/nsmb.1973
Structure of UvrA nucleotide excision repair protein in complex with modified DNA
Abstract
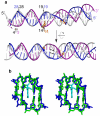
One of the primary pathways for removal of DNA damage is nucleotide excision repair (NER). In bacteria, the UvrA protein is the component of NER that locates the lesion. A notable feature of NER is its ability to act on many DNA modifications that vary in chemical structure. So far, the mechanism underlying this broad specificity has been unclear. Here, we report the first crystal structure of a UvrA protein in complex with a chemically modified oligonucleotide. The structure shows that the UvrA dimer does not contact the site of lesion directly, but rather binds the DNA regions on both sides of the modification. The DNA region harboring the modification is deformed, with the double helix bent and unwound. UvrA uses damage-induced deformations of the DNA and a less rigid structure of the modified double helix for indirect readout of the lesion.
Figures




References
-
- Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. - PubMed
-
- Sancar A, Reardon JT. Nucleotide excision repair in E. coli and man. Adv. Protein Chem. 2004;69:43–71. - PubMed
-
- Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

