Proliferation-associated POU4F2/Brn-3b transcription factor expression is regulated by oestrogen through ERα and growth factors via MAPK pathway
- PMID: 21241485
- PMCID: PMC3109571
- DOI: 10.1186/bcr2809
Proliferation-associated POU4F2/Brn-3b transcription factor expression is regulated by oestrogen through ERα and growth factors via MAPK pathway
Abstract
Introduction: In cancer cells, elevated transcription factor-related Brn-3a regulator isolated from brain cDNA (Brn-3b) transcription factor enhances proliferation in vitro and increases tumour growth in vivo whilst conferring drug resistance and migratory potential, whereas reducing Brn-3b slows growth both in vitro and in vivo. Brn-3b regulates distinct groups of key target genes that control cell growth and behaviour. Brn-3b is elevated in >65% of breast cancer biopsies, but mechanisms controlling its expression in these cells are not known.
Methods: Bioinformatics analysis was used to identify the regulatory promoter region and map transcription start site as well as transcription factor binding sites. Polymerase chain reaction (PCR) cloning was used to generate promoter constructs for reporter assays. Chromatin immunoprecipitation and site-directed mutagenesis were used to confirm the transcription start site and autoregulation. MCF-7 and Cos-7 breast cancer cells were used. Cells grown in culture were transfected with Brn-3b promoter and treated with growth factors or estradiol to test for effects on promoter activity. Quantitative reverse transcriptase PCR assays and immunoblotting were used to confirm changes in gene and protein expression.
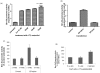
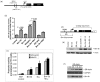
Results: We cloned the Brn-3b promoter, mapped the transcription start site and showed stimulation by estradiol and growth factors, nerve growth factor and epidermal growth factor, which are implicated in breast cancer initiation and/or progression. The effects of growth factors are mediated through the mitogen-activated protein kinase pathway, whereas hormone effects act via oestrogen receptor α (ERα). Brn-3b also autoregulates its expression and cooperates with ERα to further enhance levels.
Conclusions: Key regulators of growth in cancer cells, for example, oestrogens and growth factors, can stimulate Brn-3b expression, and autoregulation also contributes to increasing Brn-3b in breast cancers. Since increasing Brn-3b profoundly enhances growth in these cells, understanding how Brn-3b is increased in breast cancers will help to identify strategies for reducing its expression and thus its effects on target genes, thereby reversing its effects in breast cancer cells.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

