Response of an actin filament network model under cyclic stretching through a coarse grained Monte Carlo approach
- PMID: 21241710
- PMCID: PMC3501734
- DOI: 10.1016/j.jtbi.2011.01.011
Response of an actin filament network model under cyclic stretching through a coarse grained Monte Carlo approach
Abstract
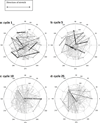
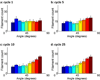
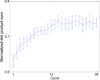
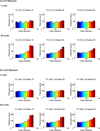
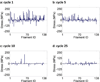
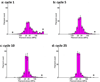
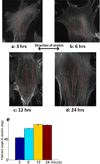
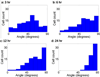
Cells are complex, dynamic systems that actively adapt to various stimuli including mechanical alterations. Central to understanding cellular response to mechanical stimulation is the organization of the cytoskeleton and its actin filament network. In this manuscript, we present a minimalistic network Monte Carlo based approach to model actin filament organization under cyclic stretching. Utilizing a coarse-grained model, a filament network is prescribed within a two-dimensional circular space through nodal connections. When cyclically stretched, the model demonstrates that a perpendicular alignment of the filaments to the direction of stretch emerges in response to nodal repositioning to minimize net nodal forces from filament stress states. In addition, the filaments in the network rearrange and redistribute themselves to reduce the overall stress by decreasing their individual stresses. In parallel, we cyclically stretch NIH 3T3 fibroblasts and find a similar cytoskeletal response. With this work, we test the hypothesis that a first-principles mechanical model of filament assembly in a confined space is by itself capable of yielding the remodeling behavior observed experimentally. Identifying minimal mechanisms sufficient to reproduce mechanical influences on cellular structure has important implications in a diversity of fields, including biology, physics, medicine, computer science, and engineering.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

References
-
- Alberts B. Molecular biology of the cell. 4th edit. New York: Garland Science; 2002.
-
- Bausch AR, Kroy K. A bottom-up approach to cell mechanics. Nat Phys. 2006;2(4):231–238.
-
- Chen CS, Alonso JL, Ostuni E, Whitesides GM, Ingber DE. Cell shape provides global control of focal adhesion assembly. Biochem Biophys Res Commun. 2003;307(2):355–361. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

