High-resolution genome-wide mapping of the primary structure of chromatin
- PMID: 21241889
- PMCID: PMC3061432
- DOI: 10.1016/j.cell.2011.01.003
High-resolution genome-wide mapping of the primary structure of chromatin
Abstract
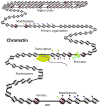
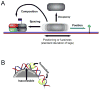
The genomic organization of chromatin is increasingly recognized as a key regulator of cell behavior, but deciphering its regulation mechanisms requires detailed knowledge of chromatin's primary structure-the assembly of nucleosomes throughout the genome. This Primer explains the principles for mapping and analyzing the primary organization of chromatin on a genomic scale. After introducing chromatin organization and its impact on gene regulation and human health, we then describe methods that detect nucleosome positioning and occupancy levels using chromatin immunoprecipitation in combination with deep sequencing (ChIP-Seq), a strategy that is now straightforward and cost efficient. We then explore current strategies for converting the sequence information into knowledge about chromatin, an exciting challenge for biologists and bioinformaticians.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




References
-
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

