Post-transcriptional control of circadian rhythms
- PMID: 21242310
- PMCID: PMC3021995
- DOI: 10.1242/jcs.065771
Post-transcriptional control of circadian rhythms
Abstract
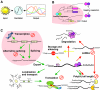
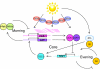
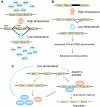
Circadian rhythms exist in most living organisms. The general molecular mechanisms that are used to generate 24-hour rhythms are conserved among organisms, although the details vary. These core clocks consist of multiple regulatory feedback loops, and must be coordinated and orchestrated appropriately for the fine-tuning of the 24-hour period. Many levels of regulation are important for the proper functioning of the circadian clock, including transcriptional, post-transcriptional and post-translational mechanisms. In recent years, new information about post-transcriptional regulation in the circadian system has been discovered. Such regulation has been shown to alter the phase and amplitude of rhythmic mRNA and protein expression in many organisms. Therefore, this Commentary will provide an overview of current knowledge of post-transcriptional regulation of the clock genes and clock-controlled genes in dinoflagellates, plants, fungi and animals. This article will also highlight how circadian gene expression is modulated by post-transcriptional mechanisms and how this is crucial for robust circadian rhythmicity.
Figures

References
-
- Baggs J. E., Green C. B. (2003). Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 13, 189-198 - PubMed
-
- Belanger V., Picard N., Cermakian N. (2006). The circadian regulation of Presenilin-2 gene expression. Chronobiol. Int. 23, 747-766 - PubMed
-
- Belostotsky D. (2009). Exosome complex and pervasive transcription in eukaryotic genomes. Curr. Opin. Cell Biol. 21, 352-358 - PubMed
-
- Brunner M., Kaldi K. (2008). Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol. Microbiol. 68, 255-262 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

