The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit
- PMID: 21242887
- PMCID: PMC3032057
- DOI: 10.1097/COH.0b013e328340ffbb
The molecular biology of HIV latency: breaking and restoring the Tat-dependent transcriptional circuit
Abstract
Purpose of review: Despite the remarkable success of intensive antiretroviral drug therapy in blocking the HIV replication, the virus persists in a small number of cells in which HIV has been transcriptionally silenced. This review will focus on recent insights into the HIV transcriptional control mechanisms that provide the biochemical basis for understanding latency.
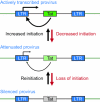
Recent findings: Latency arises when the regulatory feedback mechanism driven by HIV Tat expression is disrupted. Small changes in transcriptional initiation, induced by epigenetic silencing, can lead to restrictions in Tat levels and entry of proviruses into latency. In resting memory T-cells, which carry the bulk of the latent viral pool, additional restrictions limiting cellular levels of the essential Tat cofactor P-TEFb and the transcription initiation factors nuclear factor kappa B and nuclear factor of activated T cells ensure that the provirus remains silenced unless the host cell is activated.
Summary: Strategies to purge the latent proviral pool require nontoxic activator molecules. The multiple restrictions imposed on latent proviruses that need to be overcome suggest that proviral reactivation will not be achieved when only a single reactivation step is targeted but will require both removal of epigenetic blocks and the activation of P-TEFb. Alternatively, new inhibitors that block proviral reactivation could be developed.
Figures


Similar articles
-
Control of HIV latency by epigenetic and non-epigenetic mechanisms.Curr HIV Res. 2011 Dec 1;9(8):554-67. doi: 10.2174/157016211798998736. Curr HIV Res. 2011. PMID: 22211660 Free PMC article.
-
T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway.J Mol Biol. 2011 Jul 29;410(5):896-916. doi: 10.1016/j.jmb.2011.03.054. J Mol Biol. 2011. PMID: 21763495 Free PMC article.
-
Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure.Virology. 2014 Apr;454-455:328-39. doi: 10.1016/j.virol.2014.02.008. Epub 2014 Feb 22. Virology. 2014. PMID: 24565118 Free PMC article. Review.
-
The Molecular Basis for Human Immunodeficiency Virus Latency.Annu Rev Virol. 2017 Sep 29;4(1):261-285. doi: 10.1146/annurev-virology-101416-041646. Epub 2017 Jul 17. Annu Rev Virol. 2017. PMID: 28715973 Review.
-
The Tat Inhibitor Didehydro-Cortistatin A Prevents HIV-1 Reactivation from Latency.mBio. 2015 Jul 7;6(4):e00465. doi: 10.1128/mBio.00465-15. mBio. 2015. PMID: 26152583 Free PMC article.
Cited by
-
HIV-1 transcription and latency: an update.Retrovirology. 2013 Jun 26;10:67. doi: 10.1186/1742-4690-10-67. Retrovirology. 2013. PMID: 23803414 Free PMC article. Review.
-
Probabilistic control of HIV latency and transactivation by the Tat gene circuit.Proc Natl Acad Sci U S A. 2018 Dec 4;115(49):12453-12458. doi: 10.1073/pnas.1811195115. Epub 2018 Nov 19. Proc Natl Acad Sci U S A. 2018. PMID: 30455316 Free PMC article.
-
HIV-1 Proviral Transcription and Latency in the New Era.Viruses. 2020 May 18;12(5):555. doi: 10.3390/v12050555. Viruses. 2020. PMID: 32443452 Free PMC article. Review.
-
FACT Proteins, SUPT16H and SSRP1, Are Transcriptional Suppressors of HIV-1 and HTLV-1 That Facilitate Viral Latency.J Biol Chem. 2015 Nov 6;290(45):27297-27310. doi: 10.1074/jbc.M115.652339. Epub 2015 Sep 16. J Biol Chem. 2015. PMID: 26378236 Free PMC article.
-
Monocyte to macrophage differentiation and changes in cellular redox homeostasis promote cell type-specific HIV latency reactivation.Proc Natl Acad Sci U S A. 2024 May 7;121(19):e2313823121. doi: 10.1073/pnas.2313823121. Epub 2024 Apr 29. Proc Natl Acad Sci U S A. 2024. PMID: 38683980 Free PMC article.
References
-
- Chun TW, Davey RT, Jr., Engel D, Lane HC, Fauci AS. Re-emergence of HIV after stopping therapy. Nature. 1999;401:874–875. - PubMed
-
- Davey RT, Jr., Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc. Natl. Acad. Sci. U S A. 1999;96:15109–15114. - PMC - PubMed
-
- Shen A, Zink MC, Mankowski JL, Chadwick K, Margolick JB, Carruth LM, Li M, Clements JE, Siliciano RF. Resting CD4+ T lymphocytes but not thymocytes provide a latent viral reservoir in a simian immunodeficiency virus-Macaca nemestrina model of human immunodeficiency virus type 1-infected patients on highly active antiretroviral therapy. J. Virol. 2003;77:4938–4949. - PMC - PubMed
-
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

