A chemical method for labeling lysine methyltransferase substrates
- PMID: 21243721
- PMCID: PMC3056122
- DOI: 10.1002/cbic.201000433
A chemical method for labeling lysine methyltransferase substrates
Abstract
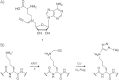
Several protein lysine methyltransferases (PKMTs) modify histones to regulate chromatin-dependent cellular processes, such as transcription, DNA replication and DNA damage repair. PKMTs are likely to have many additional substrates in addition to histones, but relatively few nonhistone substrates have been characterized, and the substrate specificity for many PKMTs has yet to be defined. Thus, new unbiased methods are needed to find PKMT substrates. Here, we describe a chemical biology approach for unbiased, proteome-wide identification of novel PKMT substrates. Our strategy makes use of an alkyne-bearing S-adenosylmethionine (SAM) analogue, which is accepted by the PKMT, SETDB1, as a cofactor, resulting in the enzymatic attachment of a terminal alkyne to its substrate. Such labeled proteins can then be treated with azide-functionalized probes to ligate affinity handles or fluorophores to the PKMT substrates. As a proof-of-concept, we have used SETDB1 to transfer the alkyne moiety from the SAM analogue onto a recombinant histone H3 substrate. We anticipate that this chemical method will find broad use in epigenetics to enable unbiased searches for new PKMT substrates by using recombinant enzymes and unnatural SAM cofactors to label and purify many substrates simultaneously from complex organelle or cell extracts.
Copyright © 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures


Similar articles
-
Expanding cofactor repertoire of protein lysine methyltransferase for substrate labeling.ACS Chem Biol. 2011 Jul 15;6(7):679-84. doi: 10.1021/cb2000567. Epub 2011 Apr 22. ACS Chem Biol. 2011. PMID: 21495674 Free PMC article.
-
Kinetic isotope effects reveal early transition state of protein lysine methyltransferase SET8.Proc Natl Acad Sci U S A. 2016 Dec 27;113(52):E8369-E8378. doi: 10.1073/pnas.1609032114. Epub 2016 Dec 9. Proc Natl Acad Sci U S A. 2016. PMID: 27940912 Free PMC article.
-
Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: participation of the chromodomain in enzymatic catalysis.Biochemistry. 2006 Mar 14;45(10):3272-84. doi: 10.1021/bi051997r. Biochemistry. 2006. PMID: 16519522
-
Protein Lysine Methyltransferases Inhibitors.Curr Med Chem. 2023;30(27):3060-3089. doi: 10.2174/0929867329666220829151257. Curr Med Chem. 2023. PMID: 36043747 Review.
-
Approaches and Guidelines for the Identification of Novel Substrates of Protein Lysine Methyltransferases.Cell Chem Biol. 2016 Sep 22;23(9):1049-1055. doi: 10.1016/j.chembiol.2016.07.013. Epub 2016 Aug 25. Cell Chem Biol. 2016. PMID: 27569752 Review.
Cited by
-
Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.Nucleic Acids Res. 2015 May 19;43(9):4602-13. doi: 10.1093/nar/gkv206. Epub 2015 Apr 8. Nucleic Acids Res. 2015. PMID: 25855808 Free PMC article.
-
Methionine Adenosyltransferase Engineering to Enable Bioorthogonal Platforms for AdoMet-Utilizing Enzymes.ACS Chem Biol. 2020 Mar 20;15(3):695-705. doi: 10.1021/acschembio.9b00943. Epub 2020 Mar 3. ACS Chem Biol. 2020. PMID: 32091873 Free PMC article.
-
Click Chemistry in Proteomic Investigations.Cell. 2020 Feb 20;180(4):605-632. doi: 10.1016/j.cell.2020.01.025. Epub 2020 Feb 13. Cell. 2020. PMID: 32059777 Free PMC article. Review.
-
Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.ACS Chem Biol. 2016 Sep 16;11(9):2484-91. doi: 10.1021/acschembio.6b00348. Epub 2016 Jul 14. ACS Chem Biol. 2016. PMID: 27351335 Free PMC article.
-
Methyltransferase-Directed Labeling of Biomolecules and its Applications.Angew Chem Int Ed Engl. 2017 May 2;56(19):5182-5200. doi: 10.1002/anie.201608625. Epub 2017 Apr 10. Angew Chem Int Ed Engl. 2017. PMID: 27943567 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

