Slow conformational dynamics in the cystoviral RNA-directed RNA polymerase P2: influence of substrate nucleotides and template RNA
- PMID: 21244027
- PMCID: PMC3059373
- DOI: 10.1021/bi101863g
Slow conformational dynamics in the cystoviral RNA-directed RNA polymerase P2: influence of substrate nucleotides and template RNA
Erratum in
- Biochemistry. 2011 May 3;50(17):3578
Abstract
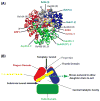
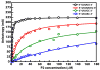
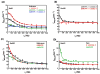
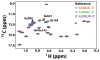
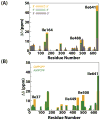
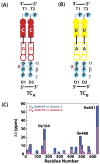
The RNA-directed RNA polymerase P2 from cystovirus ϕ6 catalyzes the de novo synthesis of positive and negative strands of the viral double-stranded RNA genome. P2 is mobile on the slow, microsecond to millisecond time scale with various motional modes, putatively assisting in RNA translocation and catalysis. Here we investigate the influence of the extreme 3'-end sequence of the single-stranded RNA templates and the nature of the substrate nucleotide triphosphates on these motional modes using multiple-quantum NMR spectroscopy. We find that P2, in the presence of templates bearing the proper genomic 3'-ends or the preferred initiation nucleotide, displays unique dynamic signatures that are different from those in the presence of nonphysiological templates or substrates. This suggests that dynamics may play a role in the fidelity of recognition of the correct substrates and template sequences to initiate RNA polymerization.
Figures
References
-
- Poranen MM, Tuma R. Self-assembly of double-stranded RNA bacteriophages. Virus Res. 2004;101:93–100. - PubMed
-
- Ferrer-Orta C, Arias A, Escarmis C, Verdaguer N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol. 2006;16:27–34. - PubMed
-
- Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

