Design of a peptide-based vector, PepFect6, for efficient delivery of siRNA in cell culture and systemically in vivo
- PMID: 21245043
- PMCID: PMC3089457
- DOI: 10.1093/nar/gkq1299
Design of a peptide-based vector, PepFect6, for efficient delivery of siRNA in cell culture and systemically in vivo
Abstract
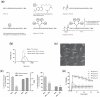
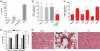
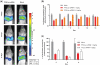
While small interfering RNAs (siRNAs) have been rapidly appreciated to silence genes, efficient and non-toxic vectors for primary cells and for systemic in vivo delivery are lacking. Several siRNA-delivery vehicles, including cell-penetrating peptides (CPPs), have been developed but their utility is often restricted by entrapment following endocytosis. Hence, developing CPPs that promote endosomal escape is a prerequisite for successful siRNA implementation. We here present a novel CPP, PepFect 6 (PF6), comprising the previously reported stearyl-TP10 peptide, having pH titratable trifluoromethylquinoline moieties covalently incorporated to facilitate endosomal release. Stable PF6/siRNA nanoparticles enter entire cell populations and rapidly promote endosomal escape, resulting in robust RNAi responses in various cell types (including primary cells), with minimal associated transcriptomic or proteomic changes. Furthermore, PF6-mediated delivery is independent of cell confluence and, in most cases, not significantly hampered by serum proteins. Finally, these nanoparticles promote strong RNAi responses in different organs following systemic delivery in mice without any associated toxicity. Strikingly, similar knockdown in liver is achieved by PF6/siRNA nanoparticles and siRNA injected by hydrodynamic infusion, a golden standard technique for liver transfection. These results imply that the peptide, in addition to having utility for RNAi screens in vitro, displays therapeutic potential.
© The Author(s) 2011. Published by Oxford University Press.
Figures




References
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- El Andaloussi S, Holm T, Langel Ü. Cell-penetrating peptides: mechanisms and applications. Curr. Pharm Des. 2005;11:3597–3611. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

