Efficient storage of high throughput DNA sequencing data using reference-based compression
- PMID: 21245279
- PMCID: PMC3083090
- DOI: 10.1101/gr.114819.110
Efficient storage of high throughput DNA sequencing data using reference-based compression
Abstract
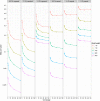
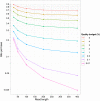
Data storage costs have become an appreciable proportion of total cost in the creation and analysis of DNA sequence data. Of particular concern is that the rate of increase in DNA sequencing is significantly outstripping the rate of increase in disk storage capacity. In this paper we present a new reference-based compression method that efficiently compresses DNA sequences for storage. Our approach works for resequencing experiments that target well-studied genomes. We align new sequences to a reference genome and then encode the differences between the new sequence and the reference genome for storage. Our compression method is most efficient when we allow controlled loss of data in the saving of quality information and unaligned sequences. With this new compression method we observe exponential efficiency gains as read lengths increase, and the magnitude of this efficiency gain can be controlled by changing the amount of quality information stored. Our compression method is tunable: The storage of quality scores and unaligned sequences may be adjusted for different experiments to conserve information or to minimize storage costs, and provides one opportunity to address the threat that increasing DNA sequence volumes will overcome our ability to store the sequences.
Figures


References
-
- Chen X, Li M, Ma B, Tromp J 2002. DNACompress: Fast and effective DNA sequence compression. Bioinformatics 18: 1696–1698 - PubMed
-
- Christley S, Lu Y, Li C, Xie X 2009. Human genomes as email attachments. Bioinformatics 25: 274–275 - PubMed
-
- Elias P 1975. Universal codeword sets and representations of the integers. IEEE Trans Inf Theory 21: 194–203
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
