SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner
- PMID: 21245377
- PMCID: PMC3067901
- DOI: 10.1128/MCB.01204-10
SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner
Abstract
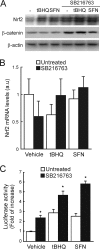
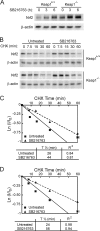
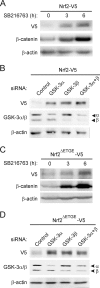
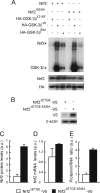
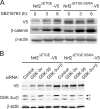
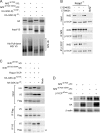
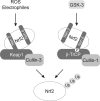
Regulation of transcription factor Nrf2 (NF-E2-related factor 2) involves redox-sensitive proteasomal degradation via the E3 ubiquitin ligase Keap1/Cul3. However, Nrf2 is controlled by other mechanisms that have not yet been elucidated. We now show that glycogen synthase kinase 3 (GSK-3) phosphorylates a group of Ser residues in the Neh6 domain of mouse Nrf2 that overlap with an SCF/β-TrCP destruction motif (DSGIS, residues 334 to 338) and promotes its degradation in a Keap1-independent manner. Nrf2 was stabilized by GSK-3 inhibitors in Keap1-null mouse embryo fibroblasts. Similarly, an Nrf2(ΔETGE) mutant, which cannot be degraded via Keap1, accumulated when GSK-3 activity was blocked. Phosphorylation of a Ser cluster in the Neh6 domain of Nrf2 stimulated its degradation because a mutant Nrf2(ΔETGE 6S/6A) protein, lacking these Ser residues, exhibited a longer half-life than Nrf2(ΔETGE). Moreover, Nrf2(ΔETGE 6S/6A) was insensitive to β-TrCP regulation and exhibited lower levels of ubiquitination than Nrf2(ΔETGE). GSK-3β enhanced ubiquitination of Nrf2(ΔETGE) but not that of Nrf2(ΔETGE 6S/6A). The Nrf2(ΔETGE) protein but not Nrf2(ΔETGE 6S/6A) coimmunoprecipitated with β-TrCP, and this association was enhanced by GSK-3β. Our results show for the first time that Nrf2 is targeted by GSK-3 for SCF/β-TrCP-dependent degradation. We propose a "dual degradation" model to describe the regulation of Nrf2 under different pathophysiological conditions.
Figures


References
-
- Apopa, P. L., X. He, and Q. Ma. 2008. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J. Biochem. Mol. Toxicol. 22:63-76. - PubMed
-
- Cuadrado, A., P. Moreno-Murciano, and J. Pedraza-Chaverri. 2009. The transcription factor Nrf2 as a new therapeutic target in Parkinson's disease. Expert Opin. Ther. Targets 13:319-329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
