Synthetic human cell fate regulation by protein-driven RNA switches
- PMID: 21245841
- PMCID: PMC3105309
- DOI: 10.1038/ncomms1157
Synthetic human cell fate regulation by protein-driven RNA switches
Abstract
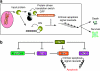
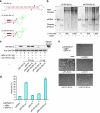
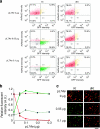
Understanding how to control cell fate is crucial in biology, medical science and engineering. In this study, we introduce a method that uses an intracellular protein as a trigger for regulating human cell fate. The ON/OFF translational switches, composed of an intracellular protein L7Ae and its binding RNA motif, regulate the expression of a desired target protein and control two distinct apoptosis pathways in target human cells. Combined use of the switches demonstrates that a specific protein can simultaneously repress and activate the translation of two different mRNAs: one protein achieves both up- and downregulation of two different proteins/pathways. A genome-encoded protein fused to L7Ae controlled apoptosis in both directions (death or survival) depending on its cellular expression. The method has potential for curing cellular defects or improving the intracellular production of useful molecules by bypassing or rewiring intrinsic signal networks.
Figures




References
-
- Keene J. D. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8, 533–543 (2007). - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

