Docosahexaenoic acid-derived neuroprotectin D1 induces neuronal survival via secretase- and PPARγ-mediated mechanisms in Alzheimer's disease models
- PMID: 21246057
- PMCID: PMC3016440
- DOI: 10.1371/journal.pone.0015816
Docosahexaenoic acid-derived neuroprotectin D1 induces neuronal survival via secretase- and PPARγ-mediated mechanisms in Alzheimer's disease models
Abstract
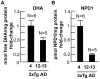
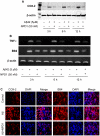
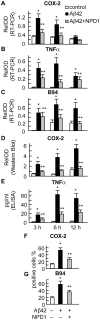
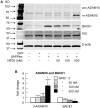
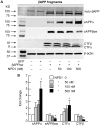
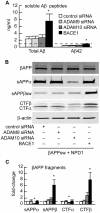
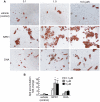
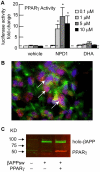
Neuroprotectin D1 (NPD1) is a stereoselective mediator derived from the omega-3 essential fatty acid docosahexaenoic acid (DHA) with potent inflammatory resolving and neuroprotective bioactivity. NPD1 reduces Aβ42 peptide release from aging human brain cells and is severely depleted in Alzheimer's disease (AD) brain. Here we further characterize the mechanism of NPD1's neurogenic actions using 3xTg-AD mouse models and human neuronal-glial (HNG) cells in primary culture, either challenged with Aβ42 oligomeric peptide, or transfected with beta amyloid precursor protein (βAPP)(sw) (Swedish double mutation APP695(sw), K595N-M596L). We also show that NPD1 downregulates Aβ42-triggered expression of the pro-inflammatory enzyme cyclooxygenase-2 (COX-2) and of B-94 (a TNF-α-inducible pro-inflammatory element) and apoptosis in HNG cells. Moreover, NPD1 suppresses Aβ42 peptide shedding by down-regulating β-secretase-1 (BACE1) while activating the α-secretase ADAM10 and up-regulating sAPPα, thus shifting the cleavage of βAPP holoenzyme from an amyloidogenic into the non-amyloidogenic pathway. Use of the thiazolidinedione peroxisome proliferator-activated receptor gamma (PPARγ) agonist rosiglitazone, the irreversible PPARγ antagonist GW9662, and overexpressing PPARγ suggests that the NPD1-mediated down-regulation of BACE1 and Aβ42 peptide release is PPARγ-dependent. In conclusion, NPD1 bioactivity potently down regulates inflammatory signaling, amyloidogenic APP cleavage and apoptosis, underscoring the potential of this lipid mediator to rescue human brain cells in early stages of neurodegenerations.
Conflict of interest statement
Figures


References
-
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. - PubMed
-
- Golde TE, Dickson D, Hutton M. Filling the gaps in the abeta cascade hypothesis of Alzheimer's disease. Curr Alzheimer Res. 2006;3:421–430. - PubMed
-
- Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. - PubMed
-
- Goedert M, Clavaguera F, Tolnay M. The propagation of prion-like protein inclusions in neurodegenerative diseases. Trends Neurosci. 2010;33:317–325. - PubMed
-
- Haass C. Initiation and propagation of neurodegeneration. Nat Med. 2010;16:1201–1204. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

