Establishment of infectious HCV virion-producing cells with newly designed full-genome replicon RNA
- PMID: 21246385
- PMCID: PMC3032175
- DOI: 10.1007/s00705-010-0859-x
Establishment of infectious HCV virion-producing cells with newly designed full-genome replicon RNA
Abstract
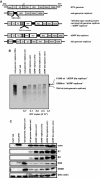
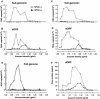
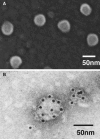
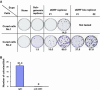
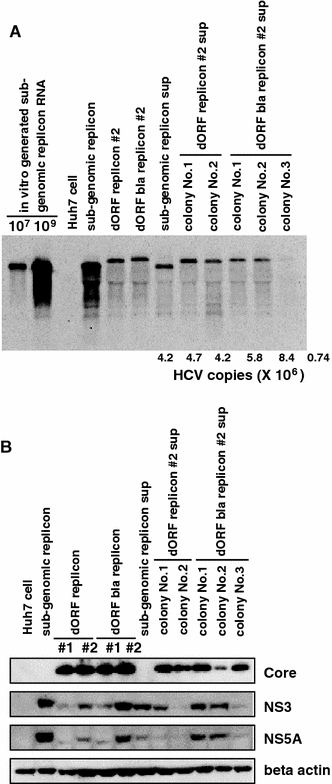
Hepatitis C virus (HCV) replicon systems enable in-depth analysis of the life cycle of HCV. However, the previously reported full-genome replicon system is unable to produce authentic virions. On the basis of these results, we constructed newly designed full-genomic replicon RNA, which is composed of the intact 5'-terminal-half RNA extending to the NS2 region flanked by an extra selection marker gene. Huh-7 cells harboring this full-genomic RNA proliferated well under G418 selection and secreted virion-like particles into the supernatant. These particles, which were round and 50 nm in diameter when analyzed by electron microscopy, had a buoyant density of 1.08 g/mL that shifted to 1.19 g/mL after NP-40 treatment; these figures match the putative densities of intact virions and nucleocapsids without envelope. The particles also showed infectivity in a colony-forming assay. This system may offer another option for investigating the life cycle of HCV.
Figures
Similar articles
-
NS2 is dispensable for efficient assembly of hepatitis C virus-like particles in a bipartite trans-encapsidation system.J Gen Virol. 2014 Nov;95(Pt 11):2427-2441. doi: 10.1099/vir.0.068932-0. Epub 2014 Jul 14. J Gen Virol. 2014. PMID: 25024280 Free PMC article.
-
An in vitro model of hepatitis C virion production.Proc Natl Acad Sci U S A. 2005 Feb 15;102(7):2579-83. doi: 10.1073/pnas.0409666102. Epub 2005 Feb 8. Proc Natl Acad Sci U S A. 2005. PMID: 15701697 Free PMC article.
-
Trans-encapsidation of hepatitis C virus subgenomic replicon RNA with viral structure proteins.Biochem Biophys Res Commun. 2008 Jul 4;371(3):446-50. doi: 10.1016/j.bbrc.2008.04.110. Epub 2008 Apr 28. Biochem Biophys Res Commun. 2008. PMID: 18445476
-
In vitro replication models for the hepatitis C virus.J Viral Hepat. 2007 Jan;14(1):2-10. doi: 10.1111/j.1365-2893.2006.00807.x. J Viral Hepat. 2007. PMID: 17212638 Review.
-
[Replication of hepatitis C virus genome].Uirusu. 2008 Dec;58(2):191-8. Uirusu. 2008. PMID: 19374197 Review. Japanese.
Cited by
-
In vivo therapeutic potential of Dicer-hunting siRNAs targeting infectious hepatitis C virus.Sci Rep. 2014 Apr 23;4:4750. doi: 10.1038/srep04750. Sci Rep. 2014. PMID: 24756133 Free PMC article.
-
Isolation and characterization of highly replicable hepatitis C virus genotype 1a strain HCV-RMT.PLoS One. 2013 Dec 16;8(12):e82527. doi: 10.1371/journal.pone.0082527. eCollection 2013. PLoS One. 2013. PMID: 24358200 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

