Comparative analysis of information contents relevant to recognition of introns in many species
- PMID: 21247441
- PMCID: PMC3033335
- DOI: 10.1186/1471-2164-12-45
Comparative analysis of information contents relevant to recognition of introns in many species
Abstract
Background: The basic process of RNA splicing is conserved among eukaryotic species. Three signals (5' and 3' splice sites and branch site) are commonly used to directly conduct splicing, while other features are also related to the recognition of an intron. Although there is experimental evidence pointing to the significant species specificities in the features of intron recognition, a quantitative evaluation of the divergence of these features among a wide variety of eukaryotes has yet to be conducted.
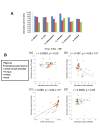
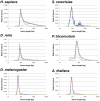
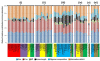
Results: To better understand the splicing process from the viewpoints of evolution and information theory, we collected introns from 61 diverse species of eukaryotes and analyzed the properties of the nucleotide sequences relevant to splicing. We found that trees individually constructed from the five features (the three signals, intron length, and nucleotide composition within an intron) roughly reflect the phylogenetic relationships among the species but sometimes extensively deviate from the species classification. The degree of topological deviation of each feature tree from the reference trees indicates the lowest discordance for the 5' splicing signal, followed by that for the 3' splicing signal, and a considerably greater discordance for the other three features. We also estimated the relative contributions of the five features to short intron recognition in each species. Again, moderate correlation was observed between the similarities in pattern of short intron recognition and the genealogical relationships among the species. When mammalian introns were categorized into three subtypes according to their terminal dinucleotide sequences, each subtype segregated into a nearly monophyletic group, regardless of the host species, with respect to the 5' and 3' splicing signals. It was also found that GC-AG introns are extraordinarily abundant in some species with high genomic G + C contents, and that the U12-type spliceosome might make a greater contribution than currently estimated in most species.
Conclusions: Overall, the present study indicates that both splicing signals themselves and their relative contributions to short intron recognition are rather susceptible to evolutionary changes, while some poorly characterized properties seem to be preserved within the mammalian intron subtypes. Our findings may afford additional clues to understanding of evolution of splicing mechanisms.
Figures






References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

