Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference
- PMID: 21247881
- PMCID: PMC3089464
- DOI: 10.1093/nar/gkq1323
Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference
Abstract
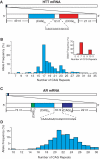
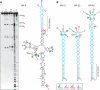
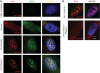
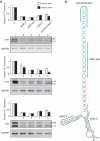
The CAG repeat expansions that occur in translated regions of specific genes can cause human genetic disorders known as polyglutamine (poly-Q)-triggered diseases. Huntington's disease and spinobulbar muscular atrophy (SBMA) are examples of these diseases in which underlying mutations are localized near other trinucleotide repeats in the huntingtin (HTT) and androgen receptor (AR) genes, respectively. Mutant proteins that contain expanded polyglutamine tracts are well-known triggers of pathogenesis in poly-Q diseases, but a toxic role for mutant transcripts has also been proposed. To gain insight into the structural features of complex triplet repeats of HTT and AR transcripts, we determined their structures in vitro and showed the contribution of neighboring repeats to CAG repeat hairpin formation. We also demonstrated that the expanded transcript is retained in the nucleus of human HD fibroblasts and is colocalized with the MBNL1 protein. This suggests that the CAG repeats in the HTT mRNA adopt ds-like RNA conformations in vivo. The intracellular structure of the CAG repeat region of mutant HTT transcripts was not sufficiently stable to be protected from cleavage by an siRNA targeting the repeats and the silencing efficiency was higher for the mutant transcript than for its normal counterpart.
© The Author(s) 2011. Published by Oxford University Press.
Figures


References
-
- Bates GP. History of genetic disease: the molecular genetics of Huntington disease - a history. Nat. Rev. Genet. 2005;6:766–773. - PubMed
-
- Martin JB, Gusella JF. Huntington's disease. Pathogenesis and management. N. Engl. J. Med. 1986;315:1267–1276. - PubMed
-
- The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. - PubMed
-
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. - PubMed
-
- Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

