The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases
- PMID: 21247896
- PMCID: PMC3060478
- DOI: 10.1074/jbc.M110.205153
The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases
Abstract
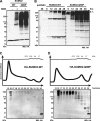
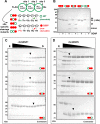
SUMOylation of proteins is a cyclic process that requires both conjugation and deconjugation of SUMO moieties. Besides modification by a single SUMO, SUMO chains have also been observed, yet the dynamics of SUMO conjugation/deconjugation remain poorly understood. Using a non-deconjugatable form of SUMO we demonstrate the underappreciated existence of SUMO chains in vivo, we highlight the importance of SUMO deconjugation, and we demonstrate the highly dynamic nature of the SUMO system. We show that SUMO-specific proteases (SENPs) play a crucial role in the dynamics of SUMO chains in vivo by constant deconjugation. Preventing deSUMOylation in Schizosaccharomyces pombe results in slow growth and a sensitivity to replication stress, highlighting the biological requirement for deSUMOylation dynamics. Furthermore, we present the mechanism of SUMO chain deconjugation by SENPs, which occurs via a stochastic mechanism, resulting in cleavage anywhere within a chain. Our results offer mechanistic insights into the workings of deSUMOylating proteases and highlight their importance in the homeostasis of (poly)SUMO-modified substrates.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

