Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes
- PMID: 21247965
- PMCID: PMC3035086
- DOI: 10.1242/dev.061416
Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes
Abstract
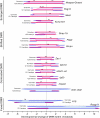
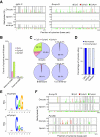
Mammalian imprinted genes are associated with differentially methylated regions (DMRs) that are CpG methylated on one of the two parental chromosomes. In mice, at least 21 DMRs acquire differential methylation in the germline and many of them act as imprint centres. We previously reported the physical extents of differential methylation at 15 DMRs in mouse embryos at 12.5 days postcoitum. To reveal the ontogeny of differential methylation, we determined and compared methylation patterns of the corresponding regions in sperm and oocytes. We found that the extent of the gametic DMRs differs significantly from that of the embryonic DMRs, especially in the case of paternal gametic DMRs. These results suggest that the gametic DMR sequences should be used to extract the features specifying methylation imprint establishment in the germline: from this analysis, we noted that the maternal gametic DMRs appear as unmethylated islands in male germ cells, which suggests a novel component in the mechanism of gamete-specific marking. Analysis of selected DMRs in blastocysts revealed dynamic changes in allelic methylation in early development, indicating that DMRs are not fully protected from the major epigenetic reprogramming events occurring during preimplantation development. Furthermore, we observed non-CpG methylation in oocytes, but not in sperm, which disappeared by the blastocyst stage. Non-CpG methylation was frequently found at maternally methylated DMRs as well as non-DMR regions, suggesting its prevalence in the oocyte genome. These results provide evidence for a unique methylation profile in oocytes and reveal the surprisingly dynamic nature of DMRs in the early embryo.
Figures




References
-
- Bourc'his D., Bestor T. H. (2004). Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96-99 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

