Coevolution of the hepatitis C virus polyprotein sites in patients on combined pegylated interferon and ribavirin therapy
- PMID: 21248044
- PMCID: PMC3067842
- DOI: 10.1128/JVI.02197-10
Coevolution of the hepatitis C virus polyprotein sites in patients on combined pegylated interferon and ribavirin therapy
Abstract
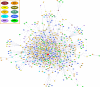
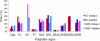
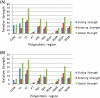
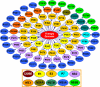
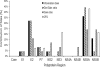
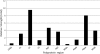
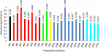
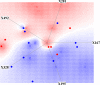
Genotype-specific sensitivity of the hepatitis C virus (HCV) to interferon-ribavirin (IFN-RBV) combination therapy and reduced HCV response to IFN-RBV as infection progresses from acute to chronic infection suggest that HCV genetic factors and intrahost HCV evolution play important roles in therapy outcomes. HCV polyprotein sequences (n = 40) from 10 patients with unsustainable response (UR) (breakthrough and relapse) and 10 patients with no response (NR) following therapy were identified through the Virahep-C study. Bayesian networks (BNs) were constructed to relate interrelationships among HCV polymorphic sites to UR/NR outcomes. All models showed an extensive interdependence of HCV sites and strong connections (P ≤ 0.003) to therapy response. Although all HCV proteins contributed to the networks, the topological properties of sites differed among proteins. E2 and NS5A together contributed ∼40% of all sites and ∼62% of all links to the polyprotein BN. The NS5A BN and E2 BN predicted UR/NR outcomes with 85% and 97.5% accuracy, respectively, in 10-fold cross-validation experiments. The NS5A model constructed using physicochemical properties of only five sites was shown to predict the UR/NR outcomes with 83.3% accuracy for 6 UR and 12 NR cases of the HALT-C study. Thus, HCV adaptation to IFN-RBV is a complex trait encoded in the interrelationships among many sites along the entire HCV polyprotein. E2 and NS5A generate broad epistatic connectivity across the HCV polyprotein and essentially shape intrahost HCV evolution toward the IFN-RBV resistance. Both proteins can be used to accurately predict the outcomes of IFN-RBV therapy.
Figures

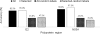
References
-
- Abid, K., R. Quadri, and F. Negro. 2000. Hepatitis C virus, the E2 envelope protein, and alpha-interferon resistance. Science 287:1555. - PubMed
-
- Andre, P., G. Perlemuter, A. Budkowska, C. Brechot, and V. Lotteau. 2005. Hepatitis C virus particles and lipoprotein metabolism. Semin. Liver Dis. 25:93-104. - PubMed
-
- Reference deleted.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

