The WNKs: atypical protein kinases with pleiotropic actions
- PMID: 21248166
- PMCID: PMC3035565
- DOI: 10.1152/physrev.00017.2010
The WNKs: atypical protein kinases with pleiotropic actions
Abstract
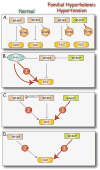
WNKs are serine/threonine kinases that comprise a unique branch of the kinome. They are so-named owing to the unusual placement of an essential catalytic lysine. WNKs have now been identified in diverse organisms. In humans and other mammals, four genes encode WNKs. WNKs are widely expressed at the message level, although data on protein expression is more limited. Soon after the WNKs were identified, mutations in genes encoding WNK1 and -4 were determined to cause the human disease familial hyperkalemic hypertension (also known as pseudohypoaldosteronism II, or Gordon's Syndrome). For this reason, a major focus of investigation has been to dissect the role of WNK kinases in renal regulation of ion transport. More recently, a different mutation in WNK1 was identified as the cause of hereditary sensory and autonomic neuropathy type II, an early-onset autosomal disease of peripheral sensory nerves. Thus the WNKs represent an important family of potential targets for the treatment of human disease, and further elucidation of their physiological actions outside of the kidney and brain is necessary. In this review, we describe the gene structure and mechanisms regulating expression and activity of the WNKs. Subsequently, we outline substrates and targets of WNKs as well as effects of WNKs on cellular physiology, both in the kidney and elsewhere. Next, consequences of these effects on integrated physiological function are outlined. Finally, we discuss the known and putative pathophysiological relevance of the WNKs.
Figures










References
-
- Abdallah JG, Schrier RW, Edelstein C, Jennings SD, Wyse B, Ellison DH. Loop diuretic infusion increases thiazide-sensitive Na(+)/Cl(−)-cotransporter abundance: role of aldosterone. J Am Soc Nephrol. 2001;12:1335–1341. - PubMed
-
- Achard JM, Warnock DG, Disse-Nicodeme S, Fiquet-Kempf B, Corvol P, Fournier A, Jeunemaitre X. Familial hyperkalemic hypertension: phenotypic analysis in a large family with the WNK1 deletion mutation. AmJMed. 2003;114:495–498. - PubMed
-
- Adragna NC, Chen Y, Delpire E, Lauf PK, Morris M. Hypertension in K-Cl cotransporter-3 knockout mice. Adv Exp Med Biol. 2004;559:379–385. - PubMed
-
- Adragna NC, Di Fulvio M, Lauf PK. Regulation of K-Cl cotransport: from function to genes. J Membr Biol. 2004;201:109–137. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

