Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription
- PMID: 21248753
- PMCID: PMC3084533
- DOI: 10.1038/nbt.1775
Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription
Abstract
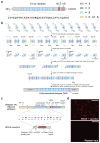
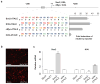
The ability to direct functional proteins to specific DNA sequences is a long-sought goal in the study and engineering of biological processes. Transcription activator-like effectors (TALEs) from Xanthomonas sp. are site-specific DNA-binding proteins that can be readily designed to target new sequences. Because TALEs contain a large number of repeat domains, it can be difficult to synthesize new variants. Here we describe a method that overcomes this problem. We leverage codon degeneracy and type IIs restriction enzymes to generate orthogonal ligation linkers between individual repeat monomers, thus allowing full-length, customized, repeat domains to be constructed by hierarchical ligation. We synthesized 17 TALEs that are customized to recognize specific DNA-binding sites, and demonstrate that they can specifically modulate transcription of endogenous genes (SOX2 and KLF4) in human cells.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Comment in
-
TALEs of genome targeting.Nat Biotechnol. 2011 Feb;29(2):135-6. doi: 10.1038/nbt.1767. Nat Biotechnol. 2011. PMID: 21301438 No abstract available.
-
TALEs for the masses.Nat Methods. 2011 Mar;8(3):197. doi: 10.1038/nmeth0311-197. Nat Methods. 2011. PMID: 21473016
-
Heritable gene targeting in zebrafish using customized TALENs.Nat Biotechnol. 2011 Aug 5;29(8):699-700. doi: 10.1038/nbt.1939. Nat Biotechnol. 2011. PMID: 21822242 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

