Autophagy in immunity and inflammation
- PMID: 21248839
- PMCID: PMC3131688
- DOI: 10.1038/nature09782
Autophagy in immunity and inflammation
Abstract
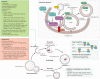
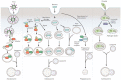
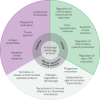
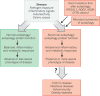
Autophagy is an essential, homeostatic process by which cells break down their own components. Perhaps the most primordial function of this lysosomal degradation pathway is adaptation to nutrient deprivation. However, in complex multicellular organisms, the core molecular machinery of autophagy - the 'autophagy proteins' - orchestrates diverse aspects of cellular and organismal responses to other dangerous stimuli such as infection. Recent developments reveal a crucial role for the autophagy pathway and proteins in immunity and inflammation. They balance the beneficial and detrimental effects of immunity and inflammation, and thereby may protect against infectious, autoimmune and inflammatory diseases.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

