Cellular SNF2H chromatin-remodeling factor promotes herpes simplex virus 1 immediate-early gene expression and replication
- PMID: 21249171
- PMCID: PMC3023163
- DOI: 10.1128/mBio.00330-10
Cellular SNF2H chromatin-remodeling factor promotes herpes simplex virus 1 immediate-early gene expression and replication
Abstract
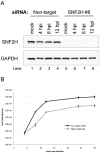
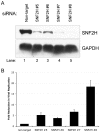
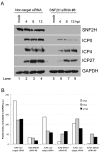
Like other DNA viruses that replicate in the nucleus, herpes simplex virus 1 (HSV-1) regulates the association of histones with its genome to promote viral replication and gene expression. We previously demonstrated that SNF2H, a member of the ISWI family of chromatin-remodeling factors, is concentrated in HSV-1 replication compartments in the nuclei of infected cells, suggesting that this cellular enzyme plays a role in viral replication. We show here that small interfering RNA (siRNA)-mediated knockdown of SNF2H in HEp-2 cells resulted in an approximately 20-fold decrease in HSV-1 replication, arguing that SNF2H promotes efficient HSV-1 replication. Decreases in HSV-1 replication were observed with multiple SNF2H-specific siRNAs, and the extent of the replication decrease correlated with the amount of SNF2H knockdown, indicating that the phenotype resulted from decreased SNF2H levels rather than off-target effects of the siRNAs. We also observed a decrease in the accumulation of immediate-early (IE) gene products in HSV-1-infected cells in which SNF2H was knocked down. Histone H3 occupancy on viral promoters was increased in HSV-1-infected cells that were transfected with SNF2H-specific siRNAs, suggesting that SNF2H promotes removal of histones from viral promoters during infection. Furthermore, chromatin immunoprecipitation (ChIP) studies showed that SNF2H associated with the HSV-1 genome during infection, which suggests that SNF2H may directly remodel viral chromatin. We hypothesize that SNF2H is recruited to viral promoters during HSV-1 infection, where it can remodel the chromatin state of the viral genome, facilitate the transcription of immediate-early genes, and enhance viral replication.
Copyright © 2011 Bryant et al
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
