Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases
- PMID: 21251613
- PMCID: PMC3229304
- DOI: 10.1016/j.ccr.2010.12.014
Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases
Abstract
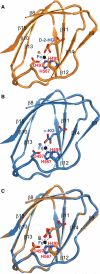
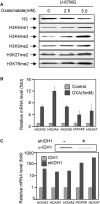
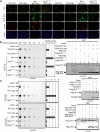
IDH1 and IDH2 mutations occur frequently in gliomas and acute myeloid leukemia, leading to simultaneous loss and gain of activities in the production of α-ketoglutarate (α-KG) and 2-hydroxyglutarate (2-HG), respectively. Here we demonstrate that 2-HG is a competitive inhibitor of multiple α-KG-dependent dioxygenases, including histone demethylases and the TET family of 5-methlycytosine (5mC) hydroxylases. 2-HG occupies the same space as α-KG does in the active site of histone demethylases. Ectopic expression of tumor-derived IDH1 and IDH2 mutants inhibits histone demethylation and 5mC hydroxylation. In glioma, IDH1 mutations are associated with increased histone methylation and decreased 5-hydroxylmethylcytosine (5hmC). Hence, tumor-derived IDH1 and IDH2 mutations reduce α-KG and accumulate an α-KG antagonist, 2-HG, leading to genome-wide histone and DNA methylation alterations.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures





References
-
- Aghili M, Zahedi F, Rafiee E. Hydroxyglutaric aciduria and malignant brain tumor: a case report and literature review. J. Neurooncol. 2009;91:233–236. - PubMed
-
- Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. - PubMed
-
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. - PubMed
-
- Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. - PubMed
-
- Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J. Inorg. Biochem. 2006;100:644–669. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

