Characterization of species C human adenovirus serotype 6 (Ad6)
- PMID: 21251688
- PMCID: PMC3056908
- DOI: 10.1016/j.virol.2010.10.041
Characterization of species C human adenovirus serotype 6 (Ad6)
Abstract
Adenovirus serotype (Ad5) is the most studied Ad. Ad1, 2, and 6 are also members of species C Ad and are presumed to have biologies similar to Ad5. In this work, we have compared the ability of Ad1, 2, 5, and 6 to infect liver and muscle after intravenous and intramuscular injection. We found that Ad6 was surprisingly the most potent at liver gene delivery and that Ad1 and Ad2 were markedly weaker than Ad5 and 6. To understand these differences, we sequenced the Ad6 genome. This revealed that the Ad6 fiber protein is surprisingly three shaft repeats shorter than the others which may explain differences in virus infectivity in vitro, but not in the liver. Comparison of hexon hypervariable regions (HVRs) suggests that the higher transduction by Ad5 and 6 as compared to Ad1 and 2 may be related to differences in charge and length.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures



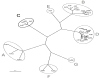
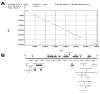




Comment in
-
Human adenovirus species C (HAdV-C) fiber protein.Virology. 2012 Mar 1;424(1):1; author reply 2. doi: 10.1016/j.virol.2011.10.017. Epub 2011 Nov 10. Virology. 2012. PMID: 22078111 No abstract available.
References
-
- Abbink P, Lemckert AA, Ewald BA, Lynch DM, Denholtz M, Smits S, Holterman L, Damen I, Vogels R, Thorner AR, O’Brien KL, Carville A, Mansfield KG, Goudsmit J, Havenga MJ, Barouch DH. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J Virol. 2007;81(9):4654–63. - PMC - PubMed
-
- Alba R, Bradshaw AC, Parker AL, Bhella D, Waddington SN, Nicklin SA, van Rooijen N, Custers J, Goudsmit J, Barouch DH, McVey JH, Baker AH. Identification of coagulation factor (F)X binding sites on the adenovirus serotype 5 hexon: effect of mutagenesis on FX interactions and gene transfer. Blood. 2009;114(5):965–71. - PMC - PubMed
-
- Capone S, Meola A, Ercole BB, Vitelli A, Pezzanera M, Ruggeri L, Davies ME, Tafi R, Santini C, Luzzago A, Fu TM, Bett A, Colloca S, Cortese R, Nicosia A, Folgori A. A novel adenovirus type 6 (Ad6)-based hepatitis C virus vector that overcomes preexisting anti-ad5 immunity and induces potent and broad cellular immune responses in rhesus macaques. J Virol. 2006;80(4):1688–99. - PMC - PubMed
-
- Croyle MA, Le HT, Linse KD, Cerullo V, Toietta G, Beaudet A, Pastore L. PEGylated helper-dependent adenoviral vectors: highly efficient vectors with an enhanced safety profile. Gene Ther. 2005;12(7):579–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

