Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants
- PMID: 21253019
- PMCID: PMC3017055
- DOI: 10.1371/journal.pone.0015853
Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants
Abstract
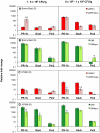
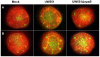
Ralstonia solanacearum, which causes bacterial wilt of diverse plants, produces copious extracellular polysaccharide (EPS), a major virulence factor. The function of EPS in wilt disease is uncertain. Leading hypotheses are that EPS physically obstructs plant water transport, or that EPS cloaks the bacterium from host plant recognition and subsequent defense. Tomato plants infected with R. solanacearum race 3 biovar 2 strain UW551 and tropical strain GMI1000 upregulated genes in both the ethylene (ET) and salicylic acid (SA) defense signal transduction pathways. The horizontally wilt-resistant tomato line Hawaii7996 activated expression of these defense genes faster and to a greater degree in response to R. solanacearum infection than did susceptible cultivar Bonny Best. However, EPS played different roles in resistant and susceptible host responses to R. solanacearum. In susceptible plants the wild-type and eps(-) mutant strains induced generally similar defense responses. But in resistant Hawaii7996 tomato plants, the wild-type pathogens induced significantly greater defense responses than the eps(-) mutants, suggesting that the resistant host recognizes R. solanacearum EPS. Consistent with this idea, purified EPS triggered significant SA pathway defense gene expression in resistant, but not in susceptible, tomato plants. In addition, the eps(-) mutant triggered noticeably less production of defense-associated reactive oxygen species in resistant tomato stems and leaves, despite attaining similar cell densities in planta. Collectively, these data suggest that bacterial wilt-resistant plants can specifically recognize EPS from R. solanacearum.
Conflict of interest statement
Figures




References
-
- Jones J, Dangl J. The plant immune system. Nature. 2006;444:323–329. - PubMed
-
- Zipfel C. Early molecular events in PAMP-triggered immunity. Curr Opin Plant Biol. 2009;12:414–420. - PubMed
-
- Strange RN, Scott PR. Plant disease: a threat to global food security. Annu Rev Phytopathol. 2005;43:83–116. - PubMed
-
- Denny T. Plant pathogenic Ralstonia species. In: Gnanamanickam SS, editor. Plant-Associated Bacteria. New York: Springer Press; 2006.
-
- Hanson PM, Wang J, Licardo O, Haudin, Mah S, et al. Variable reaction of tomato lines to bacterial wilt evaluated at several locations in southeast Asia. Hort Science. 1996;31:143–146.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

