Deciphering human heat shock transcription factor 1 regulation via post-translational modification in yeast
- PMID: 21253609
- PMCID: PMC3017095
- DOI: 10.1371/journal.pone.0015976
Deciphering human heat shock transcription factor 1 regulation via post-translational modification in yeast
Abstract
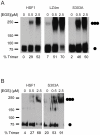
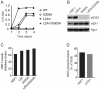
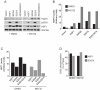
Heat shock transcription factor 1 (HSF1) plays an important role in the cellular response to proteotoxic stresses. Under normal growth conditions HSF1 is repressed as an inactive monomer in part through post-translation modifications that include protein acetylation, sumoylation and phosphorylation. Upon exposure to stress HSF1 homotrimerizes, accumulates in nucleus, binds DNA, becomes hyper-phosphorylated and activates the expression of stress response genes. While HSF1 and the mechanisms that regulate its activity have been studied for over two decades, our understanding of HSF1 regulation remains incomplete. As previous studies have shown that HSF1 and the heat shock response promoter element (HSE) are generally structurally conserved from yeast to metazoans, we have made use of the genetically tractable budding yeast as a facile assay system to further understand the mechanisms that regulate human HSF1 through phosphorylation of serine 303. We show that when human HSF1 is expressed in yeast its phosphorylation at S303 is promoted by the MAP-kinase Slt2 independent of a priming event at S307 previously believed to be a prerequisite. Furthermore, we show that phosphorylation at S303 in yeast and mammalian cells occurs independent of GSK3, the kinase primarily thought to be responsible for S303 phosphorylation. Lastly, while previous studies have suggested that S303 phosphorylation represses HSF1-dependent transactivation, we now show that S303 phosphorylation also represses HSF1 multimerization in both yeast and mammalian cells. Taken together, these studies suggest that yeast cells will be a powerful experimental tool for deciphering aspects of human HSF1 regulation by post-translational modifications.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

