Elimination of hepatitis C virus from hepatocytes by a selective activation of therapeutic molecules
- PMID: 21253612
- PMCID: PMC3017098
- DOI: 10.1371/journal.pone.0015967
Elimination of hepatitis C virus from hepatocytes by a selective activation of therapeutic molecules
Abstract
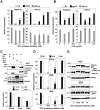
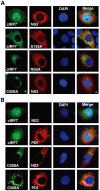
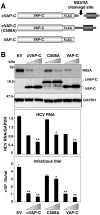
To eliminate hepatitis C virus (HCV) from infected hepatocytes, we generated two therapeutic molecules specifically activated in cells infected with HCV. A dominant active mutant of interferon (IFN) regulatory factor 7 (IRF7) and a negative regulator of HCV replication, VAP-C (Vesicle-associated membrane protein-associated protein subtype C), were fused with the C-terminal region of IPS-1 (IFNβ promoter stimulator-1), which includes an HCV protease cleavage site that was modified to be localized on the ER membrane, and designated cIRF7 and cVAP-C, respectively. In cells expressing the HCV protease, cIRF7 was cleaved and the processed fragment was migrated into the nucleus, where it activated various IFN promoters, including promoters of IFNα6, IFNβ, and IFN stimulated response element. Activation of the IFN promoters and suppression of viral RNA replication were observed in the HCV replicon cells and in cells infected with the JFH1 strain of HCV (HCVcc) by expression of cIRF7. Suppression of viral RNA replication was observed even in the IFN-resistant replicon cells by the expression of cIRF7. Expression of the cVAP-C also resulted in suppression of HCV replication in both the replicon and HCVcc infected cells. These results suggest that delivery of the therapeutic molecules into the liver of hepatitis C patients, followed by selective activation of the molecules in HCV-infected hepatocytes, is a feasible method for eliminating HCV.
Conflict of interest statement
Figures




Similar articles
-
Hepatitis C virus replication in mouse cells is restricted by IFN-dependent and -independent mechanisms.Gastroenterology. 2013 Dec;145(6):1414-23.e1. doi: 10.1053/j.gastro.2013.08.037. Epub 2013 Aug 21. Gastroenterology. 2013. PMID: 23973921
-
Restoration of the activated Rig-I pathway in hepatitis C virus (HCV) replicon cells by HCV protease, polymerase, and NS5A inhibitors in vitro at clinically relevant concentrations.Antimicrob Agents Chemother. 2013 Sep;57(9):4417-26. doi: 10.1128/AAC.00399-13. Epub 2013 Jul 8. Antimicrob Agents Chemother. 2013. PMID: 23836176 Free PMC article.
-
ME3738 enhances the effect of interferon and inhibits hepatitis C virus replication both in vitro and in vivo.J Hepatol. 2011 Jul;55(1):11-8. doi: 10.1016/j.jhep.2010.10.017. Epub 2010 Nov 29. J Hepatol. 2011. PMID: 21145867
-
Understanding the molecular mechanism(s) of hepatitis C virus (HCV) induced interferon resistance.Infect Genet Evol. 2013 Oct;19:113-9. doi: 10.1016/j.meegid.2013.06.025. Epub 2013 Jul 5. Infect Genet Evol. 2013. PMID: 23831932 Review.
-
Advances and challenges in studying hepatitis C virus in its native environment.Expert Rev Gastroenterol Hepatol. 2010 Oct;4(5):541-50. doi: 10.1586/egh.10.53. Expert Rev Gastroenterol Hepatol. 2010. PMID: 20932139 Review.
Cited by
-
Melatonin modulates the autophagic response in acute liver failure induced by the rabbit hemorrhagic disease virus.J Pineal Res. 2014 Apr;56(3):313-21. doi: 10.1111/jpi.12124. Epub 2014 Mar 2. J Pineal Res. 2014. PMID: 24499270 Free PMC article.
-
CEACAM1 Is Associated With the Suppression of Natural Killer Cell Function in Patients With Chronic Hepatitis C.Hepatol Commun. 2018 Sep 25;2(10):1247-1258. doi: 10.1002/hep4.1240. eCollection 2018 Oct. Hepatol Commun. 2018. PMID: 30288478 Free PMC article.
-
VAP Proteins - From Organelle Tethers to Pathogenic Host Interactors and Their Role in Neuronal Disease.Front Cell Dev Biol. 2022 Jun 8;10:895856. doi: 10.3389/fcell.2022.895856. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35756994 Free PMC article. Review.
-
What the VAP: The Expanded VAP Family of Proteins Interacting With FFAT and FFAT-Related Motifs for Interorganellar Contact.Contact (Thousand Oaks). 2021 May 9;4:25152564211012246. doi: 10.1177/25152564211012246. eCollection 2021 Jan 1. Contact (Thousand Oaks). 2021. PMID: 34036242 Free PMC article.
-
VAPC, an human endogenous inhibitor for hepatitis C virus (HCV) infection, is intrinsically unstructured but forms a "fuzzy complex" with HCV NS5B.PLoS One. 2012;7(7):e40341. doi: 10.1371/journal.pone.0040341. Epub 2012 Jul 17. PLoS One. 2012. PMID: 22815741 Free PMC article.
References
-
- Cerny A, Chisari FV. Pathogenesis of chronic hepatitis C: immunological features of hepatic injury and viral persistence. Hepatology. 1999;30:595–601. - PubMed
-
- Moriishi K, Matsuura Y. Mechanisms of hepatitis C virus infection. Antivir Chem Chemother. 2003;14:285–297. - PubMed
-
- Fried MW. Side effects of therapy of hepatitis C and their management. Hepatology. 2002;36:S237–244. - PubMed
-
- Sen GC. Viruses and interferons. Annu Rev Microbiol. 2001;55:255–281. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

