PAS/poly-HAMP signalling in Aer-2, a soluble haem-based sensor
- PMID: 21255112
- PMCID: PMC3078727
- DOI: 10.1111/j.1365-2958.2010.07477.x
PAS/poly-HAMP signalling in Aer-2, a soluble haem-based sensor
Abstract
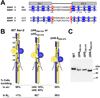
Poly-HAMP domains are widespread in bacterial chemoreceptors, but previous studies have focused on receptors with single HAMP domains. The Pseudomonas aeruginosa chemoreceptor, Aer-2, has an unusual domain architecture consisting of a PAS-sensing domain sandwiched between three N-terminal and two C-terminal HAMP domains, followed by a conserved kinase control module. The structure of the N-terminal HAMP domains was recently solved, making Aer-2 the first protein with resolved poly-HAMP structure. The role of Aer-2 in P. aeruginosa is unclear, but here we show that Aer-2 can interact with the chemotaxis system of Escherichia coli to mediate repellent responses to oxygen, carbon monoxide and nitric oxide. Using this model system to investigate signalling and poly-HAMP function, we determined that the Aer-2 PAS domain binds penta-co-ordinated b-type haem and that reversible signalling requires four of the five HAMP domains. Deleting HAMP 2 and/or 3 resulted in a kinase-off phenotype, whereas deleting HAMP 4 and/or 5 resulted in a kinase-on phenotype. Overall, these data support a model in which ligand-bound Aer-2 PAS and HAMP 2 and 3 act together to relieve inhibition of the kinase control module by HAMP 4 and 5, resulting in the kinase-on state of the Aer-2 receptor.
© 2010 Blackwell Publishing Ltd.
Figures






References
-
- Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. - PubMed
-
- Baraquet C, Theraulaz L, Iobbi-Nivol C, Mejean V, Jourlin-Castelli C. Unexpected chemoreceptors mediate energy taxis towards electron acceptors in Shewanella oneidensis. Mol Microbiol. 2009;73:278–290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

