pHg/pSILBAγ vector system for efficient gene silencing in homobasidiomycetes: optimization of ihpRNA - triggering in the mycorrhizal fungus Laccaria bicolor
- PMID: 21255319
- PMCID: PMC3836584
- DOI: 10.1111/j.1751-7915.2009.00122.x
pHg/pSILBAγ vector system for efficient gene silencing in homobasidiomycetes: optimization of ihpRNA - triggering in the mycorrhizal fungus Laccaria bicolor
Abstract
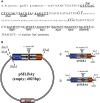
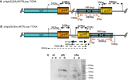
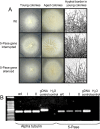
pSILBAγ silencing vector was constructed for efficient RNA silencing triggering in the model mycorrhizal fungus Laccaria bicolor. This cloning vector carries the Agaricus bisporus gpdII promoter, two multiple cloning sites separated by a L. bicolor nitrate reductase intron and the Aspergillus nidulans trpC terminator. pSILBAγ allows an easy oriented two-step PCR cloning of hairpin sequences to be expressed in basidiomycetes. With one further cloning step into pHg, a pCAMBIA1300-based binary vector carrying a hygromycin resistance cassette, the pHg/pSILBAγ plasmid is used for Agrobacterium-mediated transformation. The pHg/pSILBAγ system results in predominantly single integrations of RNA silencing triggering T-DNAs in the fungal genome and the integration sites of the transgenes can be resolved by plasmid rescue. pSILBAγ construct and two other pSILBA plasmid variants (pSILBA and pSILBAα) were evaluated for their capacity to silence Laccaria nitrate reductase gene. While all pSILBA variants tested resulted in up to 65-76% of transformants with reduced growth on nitrate, pSILBAγ produced the highest number (65%) of strongly affected fungal strains. The strongly silenced phenotype was shown to correlate with T-DNA integration in transcriptionally active genomic sites. pHg/pSILBAγ was shown to produce T-DNAs with minimum CpG methylation in transgene promoter regions which assures the maximum silencing trigger production in Laccaria. Methylation of the target endogene was only slight in RNA silencing triggered with constructs carrying an intronic spacer hairpin sequence. The silencing capacity of the pHg/pSILBAγ was further tested with Laccaria inositol-1,4,5-triphosphate 5-phosphatase gene. Besides its use in silencing triggering, the herein described plasmid system can also be used for transgene expression in Laccaria. pHg/pSILBAγ silencing system is optimized for L. bicolor but it should be highly useful also for other homobasidiomycetes, group of fungi currently lacking molecular tools for RNA silencing.
Figures





Similar articles
-
Gene knockdown by ihpRNA-triggering in the ectomycorrhizal basidiomycete fungus Laccaria bicolor.Bioeng Bugs. 2010 Sep-Oct;1(5):354-8. doi: 10.4161/bbug.1.5.12385. Bioeng Bugs. 2010. PMID: 21326837 Free PMC article.
-
RNA silencing in the model mycorrhizal fungus Laccaria bicolor: gene knock-down of nitrate reductase results in inhibition of symbiosis with Populus.Environ Microbiol. 2009 Jul;11(7):1878-96. doi: 10.1111/j.1462-2920.2009.01912.x. Epub 2009 Apr 8. Environ Microbiol. 2009. PMID: 19397683
-
T-DNA insertion, plasmid rescue and integration analysis in the model mycorrhizal fungus Laccaria bicolor.Microb Biotechnol. 2008 May;1(3):258-69. doi: 10.1111/j.1751-7915.2008.00029.x. Microb Biotechnol. 2008. PMID: 21261845 Free PMC article.
-
Agrobacterium-mediated transformation of the ectomycorrhizal symbiont Laccaria bicolor S238N.Mycorrhiza. 2005 Dec;16(1):19-22. doi: 10.1007/s00572-005-0008-7. Epub 2005 Nov 11. Mycorrhiza. 2005. PMID: 16133248
-
The Laccaria genome: a symbiont blueprint decoded.New Phytol. 2008;180(2):296-310. doi: 10.1111/j.1469-8137.2008.02613.x. New Phytol. 2008. PMID: 19138220 Review.
Cited by
-
The heat, drugs and knockout systems of microbial biotechnology.Microb Biotechnol. 2009 Nov;2(6):598-600. doi: 10.1111/j.1751-7915.2009.00144.x. Epub 2009 Aug 23. Microb Biotechnol. 2009. PMID: 21255294 Free PMC article. No abstract available.
-
Genes and evolutionary fates of the amanitin biosynthesis pathway in poisonous mushrooms.Proc Natl Acad Sci U S A. 2022 May 17;119(20):e2201113119. doi: 10.1073/pnas.2201113119. Epub 2022 May 9. Proc Natl Acad Sci U S A. 2022. PMID: 35533275 Free PMC article.
-
The development and application of a multiple gene co-silencing system using endogenous URA3 as a reporter gene in Ganoderma lucidum.PLoS One. 2012;7(8):e43737. doi: 10.1371/journal.pone.0043737. Epub 2012 Aug 24. PLoS One. 2012. PMID: 22937087 Free PMC article.
-
Fungal Shaker-like channels beyond cellular K+ homeostasis: A role in ectomycorrhizal symbiosis between Hebeloma cylindrosporum and Pinus pinaster.PLoS One. 2020 Nov 20;15(11):e0242739. doi: 10.1371/journal.pone.0242739. eCollection 2020. PLoS One. 2020. PMID: 33216794 Free PMC article.
-
Agrobacterium-mediated insertional mutagenesis in the mycorrhizal fungus Laccaria bicolor.Curr Genet. 2017 May;63(2):215-227. doi: 10.1007/s00294-016-0627-x. Epub 2016 Jul 8. Curr Genet. 2017. PMID: 27387518
References
-
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P. Genome‐wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. et al. - PubMed
-
- Audrey M.V., Bormann‐Chung C.A., Judelson H.S. Optimization of transgene‐mediated silencing in Phytophtora infestans and its association with small‐interfering RNAs. Fungal Genet Biol. 2008;45:1197–1205. - PubMed
-
- Bernstein E., Caudy A.A., Hammond S.H., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
-
- Blaise F., Remy E., Meyer M., Zhou L., Narcy J.P., Roux J. A critical assessment of Agrobacterium tumefaciens‐mediated transformation as a tool for pathogenicity gene discovery in the phytopathogenic fungus Leptosphaeria maculans. Fungal Genet Biol. 2007;44:123–138. et al. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

