Reverse transcription of the pFOXC mitochondrial retroplasmids of Fusarium oxysporum is protein primed
- PMID: 21255388
- PMCID: PMC3035579
- DOI: 10.1186/1759-8753-2-1
Reverse transcription of the pFOXC mitochondrial retroplasmids of Fusarium oxysporum is protein primed
Abstract
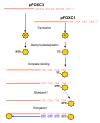
Background: The pFOXC retroplasmids are small, autonomously replicating DNA molecules found in mitochondria of certain strains of the filamentous fungus Fusarium oxysporum and are among the first linear genetic elements shown to replicate via reverse transcription. The plasmids have a unique clothespin structure that includes a 5'-linked protein and telomere-like terminal repeats, with pFOXC2 and pFOXC3 having iterative copies of a 5 bp sequence. The plasmids contain a single large open reading frame (ORF) encoding an active reverse transcriptase (RT). The pFOXC-RT is associated with the plasmid transcript in a ribonucleoprotein (RNP) complex and can synthesize full-length (-) strand cDNA products. In reactions containing partially purified RT preparations with exogenous RNAs, the pFOXC3-RT has been shown to initiate cDNA synthesis by use of snapped-back RNAs, as well as loosely associated DNA primers.
Results: The complete sequence of the distantly related pFOXC1 plasmid was determined and found to terminate in 3-5 copies of a 3 bp sequence. Unexpectedly, the majority of (-) strand cDNA molecules produced from endogenous pFOXC1 transcripts were attached to protein. In vitro experiments using partially purified pFOXC3-RT preparations having a single radiolabeled deoxyribonucleotide triphosphate (dNTP) generated a nucleotide-labeled protein that migrated at the size of the pFOXC-RT. The nucleotide preference of deoxynucleotidylation differed between pFOXC3 and pFOXC1 and showed complementarity to the respective 3' terminal repeats. In reactions that include exogenous RNA templates corresponding to the 3' end of pFOXC1, a protein-linked cDNA product was generated following deoxynucleotidylation, suggesting that reverse transcription initiates with a protein primer.
Conclusions: The finding that reverse transcription is protein primed suggests the pFOXC retroplasmids may have an evolutionary relationship with hepadnaviruses, the only other retroelement family known to initiate reverse transcription via a protein primer. Moreover, the similarity to protein-primed linear DNA elements supports models in which the terminal repeats are generated and maintained by a DNA slideback mechanism. The ability of the pFOXC-RT to utilize RNA, DNA and protein primers is unique among polymerases and suggests that the pFOXC plasmids may be evolutionary precursors of a broad range of retroelements, including hepadnaviruses, non-long terminal repeat (non-LTR) retrotransposons and telomerase.
Figures


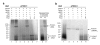



References
-
- Galligan J, Kennell J. In: Microbial Linear Plasmids. Meinhardt F, Klassen R, editor. Vol. 7. Berlin, Germany: Springer; 2007. Retroplasmids: linear and circular plasmids that replicate via reverse transcription; pp. 163–185. full_text.
-
- Eickbush TH. In: Evolutionary Biology of Viruses. Morse SS, editor. New York, USA: Raven Press; 1994. Origin and evolutionary relationships of retroelements; pp. 121–157.
LinkOut - more resources
Full Text Sources

