When the human viral infectome and diseasome networks collide: towards a systems biology platform for the aetiology of human diseases
- PMID: 21255393
- PMCID: PMC3037315
- DOI: 10.1186/1752-0509-5-13
When the human viral infectome and diseasome networks collide: towards a systems biology platform for the aetiology of human diseases
Abstract
Background: Comprehensive understanding of molecular mechanisms underlying viral infection is a major challenge towards the discovery of new antiviral drugs and susceptibility factors of human diseases. New advances in the field are expected from systems-level modelling and integration of the incessant torrent of high-throughput "-omics" data.
Results: Here, we describe the Human Infectome protein interaction Network, a novel systems virology model of a virtual virus-infected human cell concerning 110 viruses. This in silico model was applied to comprehensively explore the molecular relationships between viruses and their associated diseases. This was done by merging virus-host and host-host physical protein-protein interactomes with the set of genes essential for viral replication and involved in human genetic diseases. This systems-level approach provides strong evidence that viral proteomes target a wide range of functional and inter-connected modules of proteins as well as highly central and bridging proteins within the human interactome. The high centrality of targeted proteins was correlated to their essentiality for viruses' lifecycle, using functional genomic RNAi data. A stealth-attack of viruses on proteins bridging cellular functions was demonstrated by simulation of cellular network perturbations, a property that could be essential in the molecular aetiology of some human diseases. Networking the Human Infectome and Diseasome unravels the connectivity of viruses to a wide range of diseases and profiled molecular basis of Hepatitis C Virus-induced diseases as well as 38 new candidate genetic predisposition factors involved in type 1 diabetes mellitus.
Conclusions: The Human Infectome and Diseasome Networks described here provide a unique gateway towards the comprehensive modelling and analysis of the systems level properties associated to viral infection as well as candidate genes potentially involved in the molecular aetiology of human diseases.
Figures
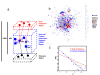
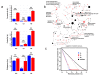
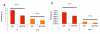
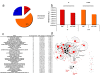


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

