The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates
- PMID: 21255825
- PMCID: PMC3065114
- DOI: 10.1016/j.cell.2011.01.001
The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates
Abstract
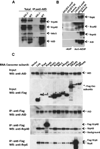
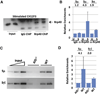
Activation-induced cytidine deaminase (AID) initiates immunoglobulin (Ig) heavy-chain (IgH) class switch recombination (CSR) and Ig variable region somatic hypermutation (SHM) in B lymphocytes by deaminating cytidines on template and nontemplate strands of transcribed DNA substrates. However, the mechanism of AID access to the template DNA strand, particularly when hybridized to a nascent RNA transcript, has been an enigma. We now implicate the RNA exosome, a cellular RNA-processing/degradation complex, in targeting AID to both DNA strands. In B lineage cells activated for CSR, the RNA exosome associates with AID, accumulates on IgH switch regions in an AID-dependent fashion, and is required for optimal CSR. Moreover, both the cellular RNA exosome complex and a recombinant RNA exosome core complex impart robust AID- and transcription-dependent DNA deamination of both strands of transcribed SHM substrates in vitro. Our findings reveal a role for noncoding RNA surveillance machinery in generating antibody diversity.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




References
-
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 2005;17:103–112. - PubMed
-
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. - PubMed
-
- Basu U, Chaudhuri J, Alpert C, Dutt S, Ranganath S, Li G, Schrum JP, Manis JP, Alt FW. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–511. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

