Dynamic local unfolding in the serpin α-1 antitrypsin provides a mechanism for loop insertion and polymerization
- PMID: 21258324
- PMCID: PMC3074950
- DOI: 10.1038/nsmb.1976
Dynamic local unfolding in the serpin α-1 antitrypsin provides a mechanism for loop insertion and polymerization
Abstract
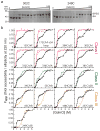
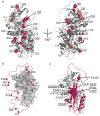
The conformational plasticity of serine protease inhibitors (serpins) underlies both their activities as protease inhibitors and their susceptibility to pathogenic misfolding and aggregation. Here, we structurally characterize a sheet-opened state of the serpin α-1 antitrypsin (α₁AT) and show how local unfolding allows functionally essential strand insertion. Mutations in α₁AT that cause polymerization-induced serpinopathies map to the labile region, suggesting that the evolution of serpin function required sampling of high risk conformations on a dynamic energy landscape.
Figures
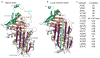
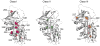

Similar articles
-
Defining the mechanism of polymerization in the serpinopathies.Proc Natl Acad Sci U S A. 2010 Oct 5;107(40):17146-51. doi: 10.1073/pnas.1004785107. Epub 2010 Sep 20. Proc Natl Acad Sci U S A. 2010. PMID: 20855577 Free PMC article.
-
The structural basis of serpin polymerization studied by hydrogen/deuterium exchange and mass spectrometry.J Biol Chem. 2008 Nov 7;283(45):30804-11. doi: 10.1074/jbc.M804048200. Epub 2008 Sep 15. J Biol Chem. 2008. PMID: 18794298 Free PMC article.
-
Probing the local conformational change of alpha1-antitrypsin.Protein Sci. 2007 Sep;16(9):1842-50. doi: 10.1110/ps.072911607. Epub 2007 Jul 27. Protein Sci. 2007. PMID: 17660256 Free PMC article.
-
Conformational changes in serpins and the mechanism of alpha 1-antitrypsin deficiency.Am J Respir Crit Care Med. 1994 Dec;150(6 Pt 2):S171-5. doi: 10.1164/ajrccm/150.6_Pt_2.S171. Am J Respir Crit Care Med. 1994. PMID: 7952655 Review.
-
New insights into the structural basis of alpha 1-antitrypsin deficiency.QJM. 1996 Nov;89(11):807-12. doi: 10.1093/qjmed/89.11.807. QJM. 1996. PMID: 8977959 Review.
Cited by
-
How the serpin α1-proteinase inhibitor folds.J Biol Chem. 2012 Apr 6;287(15):12425-32. doi: 10.1074/jbc.M111.315465. Epub 2012 Feb 13. J Biol Chem. 2012. PMID: 22334651 Free PMC article.
-
Collapse of a long axis: single-molecule Förster resonance energy transfer and serpin equilibrium unfolding.Biochemistry. 2014 May 13;53(18):2903-14. doi: 10.1021/bi401622n. Epub 2014 May 1. Biochemistry. 2014. PMID: 24749911 Free PMC article.
-
The Z mutation alters the global structural dynamics of α1-antitrypsin.PLoS One. 2014 Sep 2;9(9):e102617. doi: 10.1371/journal.pone.0102617. eCollection 2014. PLoS One. 2014. PMID: 25181470 Free PMC article.
-
Challenges and Prospects for Alpha-1 Antitrypsin Deficiency Gene Therapy.Hum Gene Ther. 2015 Nov;26(11):709-18. doi: 10.1089/hum.2015.044. Epub 2015 Sep 29. Hum Gene Ther. 2015. PMID: 26413996 Free PMC article. Review.
-
ER chaperones use a protein folding and quality control glyco-code.Mol Cell. 2023 Dec 21;83(24):4524-4537.e5. doi: 10.1016/j.molcel.2023.11.006. Epub 2023 Dec 4. Mol Cell. 2023. PMID: 38052210 Free PMC article.
References
-
- Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. - PubMed
-
- Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. - PubMed
-
- Dupont DM, et al. Biochemical properties of plasminogen activator inhibitor-1. Front Biosci. 2009;14:1337–1361. - PubMed
-
- Mushunje A, Evans G, Brennan SO, Carrell RW, Zhou A. Latent antithrombin and its detection, formation and turnover in the circulation. J Thromb Haemost. 2004;2:2170–2177. - PubMed
-
- Kaslik G, et al. Effects of serpin binding on the target proteinase: global stabilization, localized increased structural flexibility, and conserved hydrogen bonding at the active site. Biochemistry. 1997;36:5455–5464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

