PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1
- PMID: 21258370
- PMCID: PMC3077550
- DOI: 10.1038/ncb2151
PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1
Abstract
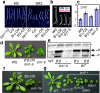
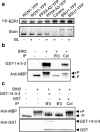
When brassinosteroid levels are low, the GSK3-like kinase BIN2 phosphorylates and inactivates the BZR1 transcription factor to inhibit growth in plants. Brassinosteroid promotes growth by inducing dephosphorylation of BZR1, but the phosphatase that dephosphorylates BZR1 has remained unknown. Here, using tandem affinity purification, we identified protein phosphatase 2A (PP2A) as a BZR1-interacting protein. Genetic analyses demonstrated a positive role for PP2A in brassinosteroid signalling and BZR1 dephosphorylation. Members of the B' regulatory subunits of PP2A directly interact with BZR1's putative PEST domain containing the site of the bzr1-1D mutation. Interaction with and dephosphorylation by PP2A are enhanced by the bzr1-1D mutation, reduced by two intragenic bzr1-1D suppressor mutations, and abolished by deletion of the PEST domain. This study reveals a crucial function for PP2A in dephosphorylating and activating BZR1 and completes the set of core components of the brassinosteroid-signalling cascade from cell surface receptor kinase to gene regulation in the nucleus.
Figures



References
-
- Kim TW, Wang ZY. Brassinosteroid Signal Transduction from Receptor Kinases to Transcription Factors. Annu Rev Plant Biol. 2010;61:681–704. - PubMed
-
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. - PubMed
-
- Clouse SD. Brassinosteroids. Plant counterparts to animal steroid hormones? Vitam Horm. 2002;65:195–223. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

