Cupredoxins--a study of how proteins may evolve to use metals for bioenergetic processes
- PMID: 21258692
- PMCID: PMC6916721
- DOI: 10.1039/c0mt00061b
Cupredoxins--a study of how proteins may evolve to use metals for bioenergetic processes
Abstract
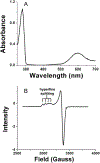
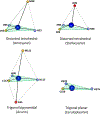
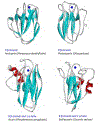
Cupredoxins are small proteins that contain type I copper centers, which are ubiquitous in nature. They function as electron transfer shuttles between proteins. This review of the structure and properties of native cupredoxins, and those modified by site-directed mutagenesis, illustrates how these proteins may have evolved to specifically bind copper, develop recognition sites for specific redox partners, tune redox potential for a particular function, and allow for efficient electron transfer through the protein matrix. This is relevant to the general understanding of the roles of metals in energy metabolism, respiration and photosynthesis.
Figures




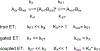
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

