RNA and protein 3D structure modeling: similarities and differences
- PMID: 21258831
- PMCID: PMC3168752
- DOI: 10.1007/s00894-010-0951-x
RNA and protein 3D structure modeling: similarities and differences
Abstract
In analogy to proteins, the function of RNA depends on its structure and dynamics, which are encoded in the linear sequence. While there are numerous methods for computational prediction of protein 3D structure from sequence, there have been very few such methods for RNA. This review discusses template-based and template-free approaches for macromolecular structure prediction, with special emphasis on comparison between the already tried-and-tested methods for protein structure modeling and the very recently developed "protein-like" modeling methods for RNA. We highlight analogies between many successful methods for modeling of these two types of biological macromolecules and argue that RNA 3D structure can be modeled using "protein-like" methodology. We also highlight the areas where the differences between RNA and proteins require the development of RNA-specific solutions.
Figures
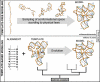
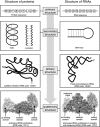
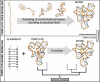
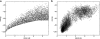
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

