Microbial whole-cell arrays
- PMID: 21261831
- PMCID: PMC3864447
- DOI: 10.1111/j.1751-7915.2007.00021.x
Microbial whole-cell arrays
Abstract
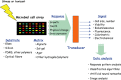
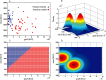
The coming of age of whole-cell biosensors, combined with the continuing advances in array technologies, has prepared the ground for the next step in the evolution of both disciplines - the whole-cell array. In the present review, we highlight the state-of-the-art in the different disciplines essential for a functional bacterial array. These include the genetic engineering of the biological components, their immobilization in different polymers, technologies for live cell deposition and patterning on different types of solid surfaces, and cellular viability maintenance. Also reviewed are the types of signals emitted by the reporter cell arrays, some of the transduction methodologies for reading these signals and the mathematical approaches proposed for their analysis. Finally, we review some of the potential applications for bacterial cell arrays, and list the future needs for their maturation: a richer arsenal of high-performance reporter strains, better methodologies for their incorporation into hardware platforms, design of appropriate detection circuits, the continuing development of dedicated algorithms for multiplex signal analysis and - most importantly - enhanced long-term maintenance of viability and activity on the fabricated biochips.
© 2008 The Authors.
Figures
References
-
- Ahn J.M., Mitchell R.J., Gu M.B. Detection and classification of oxidative damaging stresses using recombinant bioluminescent bacteria harboring sodapqi::, and katgluxCDABE fusions. Enzyme Microb Technol. 2004;35:540–544.
-
- Albrecht D.R., Tsang V.L., Sah R.L., Bhatia S.N. Photo‐ and electropatterning of hydrogel‐encapsulated living cell arrays. Lab Chip. 2005;5:111–118. - PubMed
-
- Alper J. Biology and the inkjets. Science. 2004;305:1895. - PubMed
-
- Aravanis A.M., DeBusschere B.D., Chruscinski A.J., Gilchrist K.H., Kobilka B.K., Kovacs G.T.A. A genetically engineered cell‐based biosensor for functional classification of agents. Biosens Bioelectron. 2001;16:571–577. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

