Decreasing Enterobacter sakazakii (Cronobacter spp.) food contamination level with bacteriophages: prospects and problems
- PMID: 21261874
- PMCID: PMC3815295
- DOI: 10.1111/j.1751-7915.2008.00058.x
Decreasing Enterobacter sakazakii (Cronobacter spp.) food contamination level with bacteriophages: prospects and problems
Abstract
Enterobacter sakazakii (Cronobacter spp.) is an opportunistic pathogen, which can cause rare, but life-threatening infections in neonates and infants through feeding of a contaminated milk formula. We isolated 67 phages from environmental samples and tested their lytic host range on a representative collection of 40 E. sakazakii strains. A cocktail of five phages prevented the outgrowth of 35 out of 40 test strains in artificially contaminated infant formula. Two E. sakazakii phages represented prolate head Myoviridae. Molecular tests identified them as close relatives of Escherichia coli phage T4. The remaining three phages represented isometric head Myoviridae with large genome size of 140 and 200 kb, respectively, which belonged to two different DNA hybridization groups. A high dose of 10(8) pfu ml(-1) of phage could effectively sterilize a broth contaminated with both high and low pathogen counts (10(6) and 10(2) cfu ml(-1)). In contrast, broth inoculated with 10(4) phage and 10(2) bacteria per ml first showed normal bacterial growth until reaching a cell titre of 10(5) cfu ml(-1). Only when crossing this threshold, phage replication started, but it could not reduce the contamination level below 100 cfu ml(-1). Phages could be produced with titres of 10(10) pfu ml(-1) in broth culture, but they were not stable upon freeze-drying. Addition of trehalose or milk formula stabilized the phage preparation, which then showed excellent storage stability even at elevated temperature.
© 2008 The Authors. Journal compilation © 2008 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures


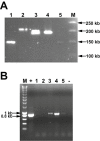
 concatemers, 50 kb DNA ladder, Promega).
B. PCR analysis on phage DNA with primers Mzia1 and CAP8 amplifying the major head gene g23 (Tétart et al., 2001). (M), Smartladder (Eurogentec); (+), T4 positive control; (1), phage F; (2), phage 23; (3), phage 81; (4), phage 82; (5), phage 83; and (−), negative control without DNA.
concatemers, 50 kb DNA ladder, Promega).
B. PCR analysis on phage DNA with primers Mzia1 and CAP8 amplifying the major head gene g23 (Tétart et al., 2001). (M), Smartladder (Eurogentec); (+), T4 positive control; (1), phage F; (2), phage 23; (3), phage 81; (4), phage 82; (5), phage 83; and (−), negative control without DNA.
Similar articles
-
Investigating the biocontrol and anti-biofilm potential of a three phage cocktail against Cronobacter sakazakii in different brands of infant formula.Int J Food Microbiol. 2017 Jul 17;253:1-11. doi: 10.1016/j.ijfoodmicro.2017.04.009. Epub 2017 Apr 21. Int J Food Microbiol. 2017. PMID: 28460269
-
A Polyvalent Broad-Spectrum Escherichia Phage Tequatrovirus EP01 Capable of Controlling Salmonella and Escherichia coli Contamination in Foods.Viruses. 2022 Jan 29;14(2):286. doi: 10.3390/v14020286. Viruses. 2022. PMID: 35215879 Free PMC article.
-
A Novel Bacteriophage Targeting Cronobacter sakazakii Is a Potential Biocontrol Agent in Foods.Appl Environ Microbiol. 2015 Oct 23;82(1):192-201. doi: 10.1128/AEM.01827-15. Print 2016 Jan 1. Appl Environ Microbiol. 2015. PMID: 26497465 Free PMC article.
-
Cronobacter species (formerly known as Enterobacter sakazakii) in powdered infant formula: a review of our current understanding of the biology of this bacterium.J Appl Microbiol. 2012 Jul;113(1):1-15. doi: 10.1111/j.1365-2672.2012.05281.x. Epub 2012 Apr 11. J Appl Microbiol. 2012. PMID: 22420458 Review.
-
Nonthermal Inactivation of Cronobacter sakazakii in Infant Formula Milk: A Review.Crit Rev Food Sci Nutr. 2016 Jul 26;56(10):1620-9. doi: 10.1080/10408398.2013.781991. Crit Rev Food Sci Nutr. 2016. PMID: 25603362 Review.
Cited by
-
Bacteriophage biocontrol of foodborne pathogens.J Food Sci Technol. 2016 Mar;53(3):1355-62. doi: 10.1007/s13197-015-1996-8. Epub 2015 Oct 26. J Food Sci Technol. 2016. PMID: 27570260 Free PMC article. Review.
-
Cronobacter sakazakii: stress survival and virulence potential in an opportunistic foodborne pathogen.Gut Microbes. 2014;5(6):711-8. doi: 10.4161/19490976.2014.983774. Gut Microbes. 2014. PMID: 25562731 Free PMC article. Review.
-
Genome sequence and characterization of the Tsukamurella bacteriophage TPA2.Appl Environ Microbiol. 2011 Feb;77(4):1389-98. doi: 10.1128/AEM.01938-10. Epub 2010 Dec 23. Appl Environ Microbiol. 2011. PMID: 21183635 Free PMC article.
-
Bacteriophage Applications for Food Production and Processing.Viruses. 2018 Apr 19;10(4):205. doi: 10.3390/v10040205. Viruses. 2018. PMID: 29671810 Free PMC article. Review.
-
Complete genome sequence of Cronobacter sakazakii temperate bacteriophage phiES15.J Virol. 2012 Jul;86(14):7713-4. doi: 10.1128/JVI.01042-12. J Virol. 2012. PMID: 22733879 Free PMC article.
References
-
- Bruce J. Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 1996;50:77–81.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

